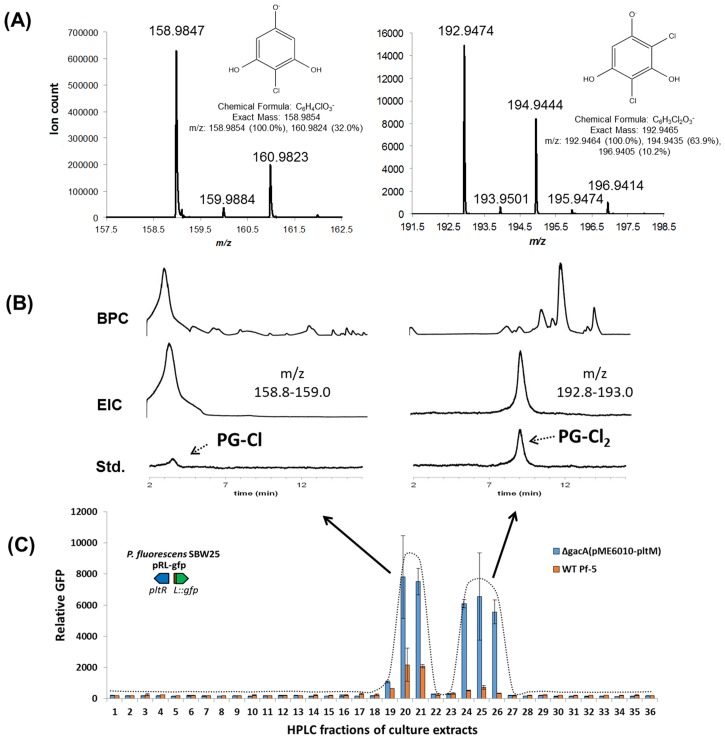
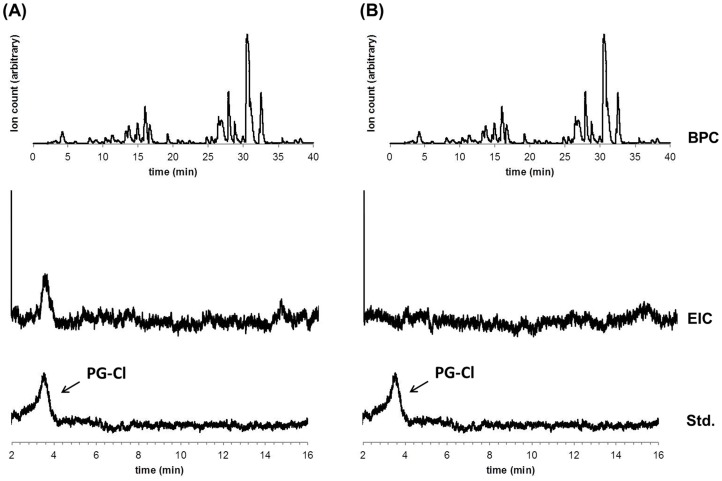
Figure 4. Identification of the signaling metabolites from Pf-5 that induce expression of pltL::gfp in P. fluorescens SBW25 (pRL-gfp).
Metabolites were extracted from wild-type (WT) Pf-5, and a phloroglucinol (PG)-fed ΔgacA mutant of Pf-5 carrying a plasmid that constitutively expresses pltM (pME6010-pltM). The extracts were fractioned into 48 fractions using semi-preparatory HPLC (no observable signal activity was detected from the last 12 fractions, data not shown). The signal activity of the fractions from both strains was tested using strain P. fluorescens SBW25 containing pRL-gfp (C). Data are means of three biological replicates, and error bars represent the standard deviation of the mean. (A) Mass spectra of compound eluting at 3.3 min in Fraction 21 (left) and 9 min in Fraction 26 (right) from the ΔgacA (pME6010-pltM) culture extracts. (B) Comparisons between the signal compounds identified in Pf-5 cultures to synthesized PG-Cl and PG-Cl2, which elute at 3.3 and 9 min, respectively, as indicated by the dashed arrows. BPC, base peak chromatogram; EIC, extracted ion chromatogram; Std, synthesized standards. The m/z used for EIC analysis of PG-Cl and PG-Cl2 are also shown.