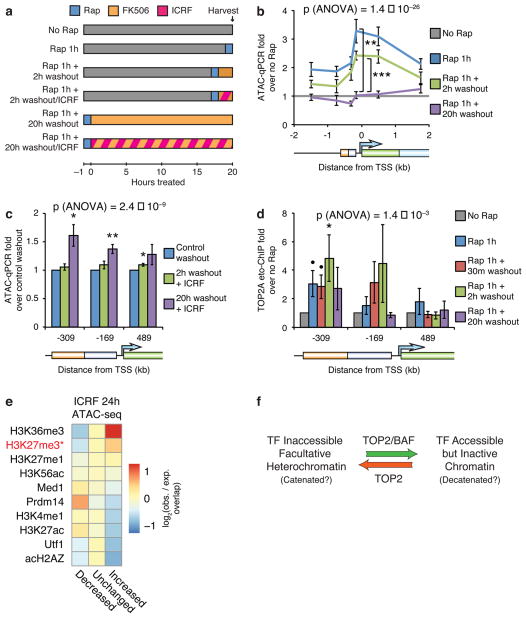
Figure 6. TOP2 is required for reformation of facultative heterochromatin.
(a) Strategy for BAF recruitment and rapamycin washout using 100 nM FK506 with TOP2 inhibition using 1 μM ICRF-193. ATAC-qPCR in fibroblasts treated with 3 nM rapamycin for 1 hour and subsequently washed out with FK506 (b) in the presence of ICRF-193 (c). (d) TOP2A etoposide-ChIP in cells treated with rapamycin and subsequently washed out with FK506. (e) Heatmap of log2 observed/expected change in accessibility of various ChIP-seq peaks, sorted by enrichment for increased accessibility upon ICRF-193 treatment. Top 10 ChIP-seq datasets are shown. *: does not include bivalent peaks. (f) Model for the role of TOP2 and BAF in resolution and reformation of heterochromatin. Significance of individual primer sets assessed by t-tests as specified (b), versus control washout (c), or versus no rapamycin control (d). Overall significance of the effect of washout (b,d) or ICRF-193 (c) assessed by three-way ANOVA. Lines and bars represent means and error bars represent s.e.m. from 5 (b,c) or 9 (d) cell passages.