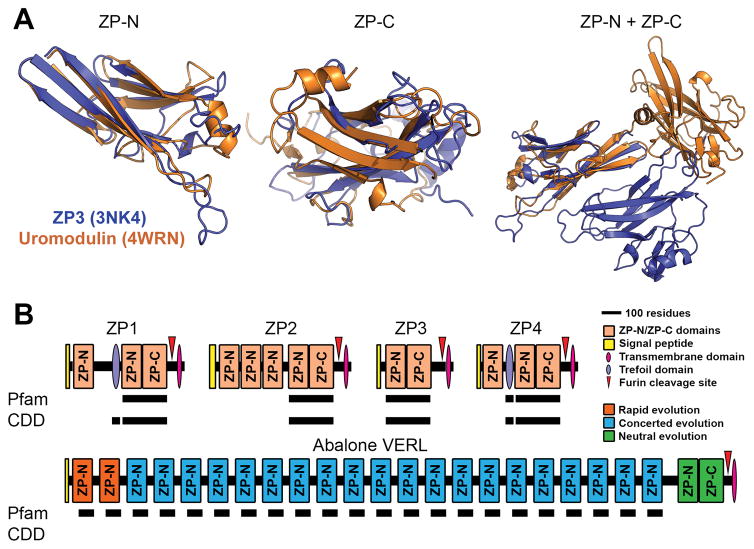
Figure 1. Protein and gene structures of ZP-N/ZP-C domain containing proteins.
(A) Comparison of chicken ZP3 (Protein Data Bank: 3NK4) and human uromodulin (4WRN). Independent alignment of the ZP-N and ZP-C domain reveal close structural similarity of ~3.4 Å and 4.2 Å rmsd, respectively. When viewed together (and aligned based on the ZP-N domain), however, there is no similarity in the positioning of ZP-C relative to ZP-N (adapted from Bokhove et al., 2016). (B) Several classical “ZP domain”-containing proteins often contain N-terminal repeats of the ZP-N domain. Structural databases such as Pfam and the Conserved Domain Database (CDD), however, do not detect these additional repeats in human ZP proteins; Pfam also did not detect the conserved trefoil domain in ZP1. The most extreme case of this ZP-N duplication is in abalone VERL, which has 22 ZP-N repeats with different modes of evolution based on genetic effects and adaptation with coevolving sperm proteins. Pfam includes an additional entry for 78 residues of the VERL repeats (pfam11386), but this annotation does not fully span the repeats or report ZP-N homology. The C-terminal ZP module of VERL is also not detected in either database (adapted from Wilburn and Swanson, 2016).