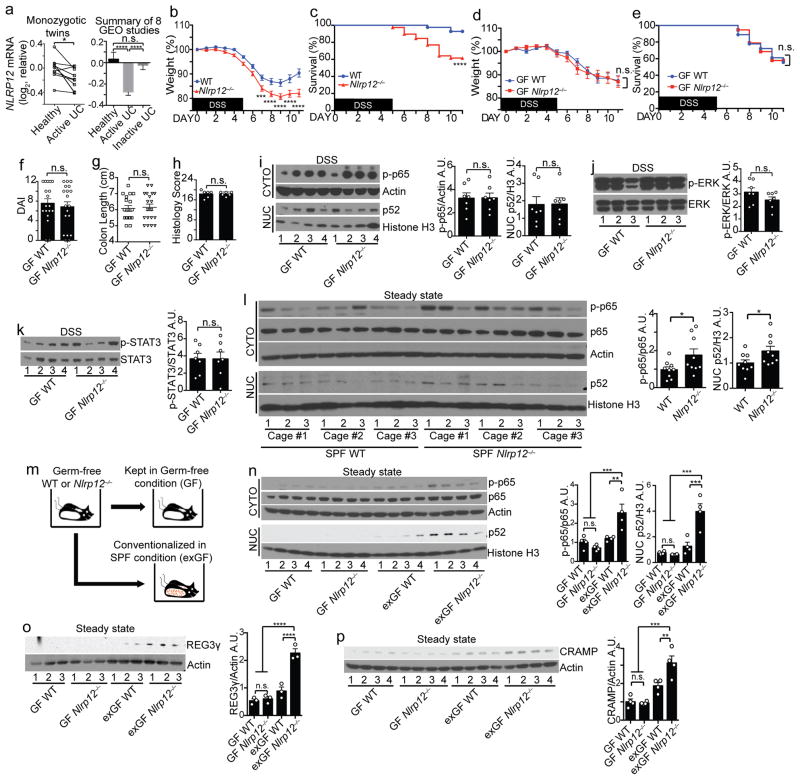
Figure 1.
Microbiota differentiates colitis severity between WT and Nlrp12−/− mice. (a) NLRP12 gene expression between 10 pairs of monozygotic healthy and UC twins (left panel). Composite of 8 NCBI GEO ulcerative colitis (UC) studies (right panel) (Supplementary Fig. 1 shows the other seven individual studies). (b) Body weight and (c) percent survival of conventionally-raised mice treated with 3% DSS (WT, n=41; Nlrp12−/−, n=42), compiled from 4 independent experiments. (d) Body weight, (e) percent survival, (f) disease-associated index (DAI) and (g) colon length of germ-free (GF) WT (n=18) and Nlrp12−/− (n=19) mice given 1.5% DSS, compiled from 2 independent experiments. (h) Blinded histopathology scoring of colons (n=6/group). (i–k) Representative immunoblots and densitometry of distal colon proteins from DSS-treated GF mice from 2 independent experiments (n=7/group). (l) Immunoblots and densitometry of distal colon cytosolic (CYTO) and nuclear (NUC) protein fractions from untreated specific-pathogen free (SPF) mice (n=9/group). (m) Schematic comparing GF and conventionalized (exGF) mice. (n–p) Immunoblots and densitometry of distal colon proteins from untreated GF and exGF mice (n,p, n=4/group; o, n=3/group). One dot or one lane represents one mouse. Error bars show SEM. *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001, and n.s. means no significance by two-tailed paired t test (a, left panel), unpaired t test (a, right panel, b, d, f–l, n–p) or Log-rank (Mantel Cox) test (c and e).