FIG 2 .
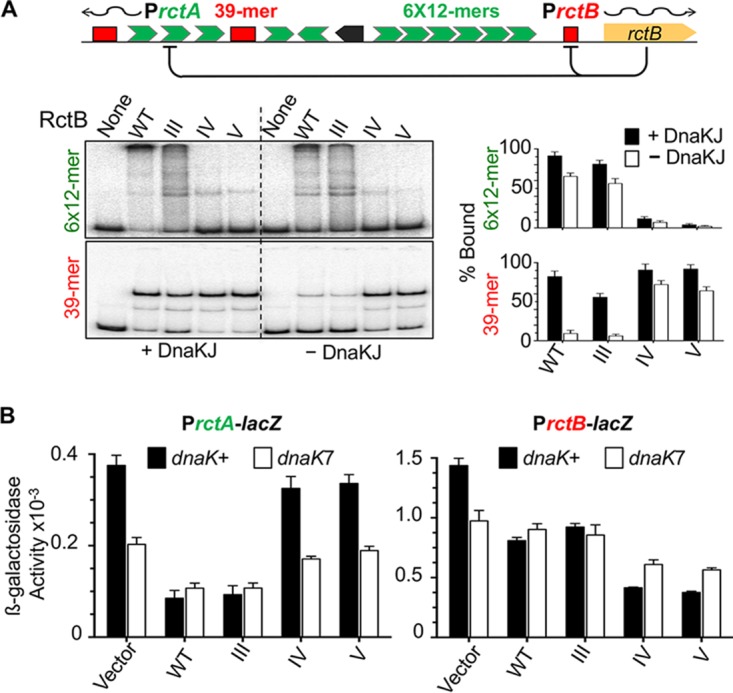
DNA binding activities of alanine mutants III to V in vitro and in vivo. (A) DNA binding of RctB determined by EMSA. The DNA fragment carried either the array of six 12-mer sites (6×12-mer) or a 39-mer site as identified in the schematic map of Chr2 origin. The fragments had 55 bp of vector sequences at both flanks of the sites. RctB, pretreated with chaperones DnaKJ (63) or not (black and white bars, respectively), was reacted with DNA at fixed concentrations (~15 nM and 1 nM, respectively) in all cases. The autoradiograms were quantified for the percentage of fragments bound by RctB, and the values are shown as bar diagrams. The solid bars represent mean levels of binding from three repeat experiments and the error bars 1 standard deviation of the mean (here and elsewhere). (B) RctB binding to 12-mers and 39-mers in vivo using a promoter repression assay. The origin of Chr2 has two promoters, PrctA and PrctB, that have overlapping 12-mer and 39-mer sites, respectively, which are repressed in the presence of RctB (origin diagram in panel A). The activities of the promoters were measured after fusing them to a promoter-less lacZ gene and measuring β-galactosidase activity. RctB proteins were same as those described for panel A, except that they were left untagged and expressed from a constitutive promoter. The vector carried the same promoter but no rctB gene. The β-galactosidase activities in dnaK+ and dnaK7 hosts are represented by black and white bars, respectively. The activities were determined from three biological replicates. The level of RctB expression was monitored by Western blotting (Fig. S2A in the supplemental material).
