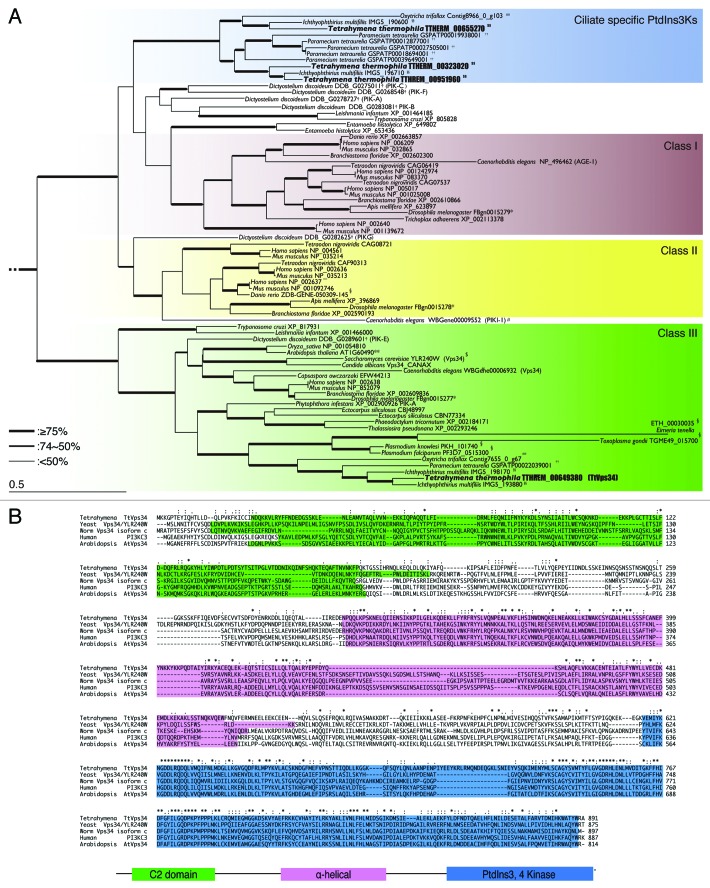
Figure 1. Characterization of phosphatidylinositol 3-kinases in Tetrahymena. (A) Phylogenetic tree of PtdIns3Ks. The tree was reconstructed with a maximum likelihood method (see Materials and Methods). Superscripts on the accession numbers stand for databases, which the protein sequences were taken from;†: dictybase, **: Tair, $: Saccharomyces Genome Database, #: WormBase, *:FlyBase, §: GeneDB, ##: OxyDB, ††: ParameciumDB, §§: IchDB, and $$:TGB. The accession numbers without superscript indicate the protein sequences that were taken from GenBank. Branches with different widths represent bootstrap values. Scale bar: 0.5, expected amino acid residue substitutions per site. (B) Multiple-sequence alignment of the whole amino acid sequence of class III PtdIns3Ks (human PIK3C3 or yeast Vps34) including Homo sapiens, Saccharomyces cerevisiae, Dictyostelium discoideum, Arabidopsis thaliana, and Tetrahymena thermophila. Each color box represents the conserved domain, which corresponds to the schematic representation of the primary structure of orthologs of yeast Vps34. Asterisks indicate identical amino acids. Colons and semicolons indicate amino acid similarity.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.