Abstract
Introduction:
Bladder cancer (BC) has varied clinical behavior in terms of recurrence and progression. Current pathological characteristics are insufficient to prognosticate the outcome of a given treatment. Cellular metabolic regulatory molecules, such as micro RNA (miRNA), could be a potential biomarker to prognosticate the treatment outcomes.
Materials and Methods:
PubMed and Google Scholar databases were searched for publications from 1990 to 2016, related to miRNA biogenesis, its function, and role in the pathogenesis of bladder as well as other cancers. Articles were searched using MeSH terms micrornas, micrornas AND neoplasm, and micrornas AND urinary bladder neoplasm. Out of the 108 publications reviewed 75 references were selected based on the clinical relevance. Articles were reviewed to assess the role of miRNA in various cancers and those in BC as a diagnostic or therapeutic tool.
Results:
More than 35 miRNAs were found to be associated with different pathways of cellular dedifferentiation, proliferation, and progression of BC as well as other cancers. A normal looking mucosa may show molecular changes preceding phenotypic changes in the form of varied expression of miR-129, miR-200a, and miR-205. miR-214, miR-99a, and miR-125b have been shown to be potential urinary biomarkers of BC. miRNAs could act as a repressor for protein molecule functioning or activator of different pathways to be used as a therapeutic target too.
Conclusions:
Despite certain limitations, such as instability, rapid plasma clearance, and targeting antagonist proteins of cellular metabolic pathways, miRNAs have potential to be studied as a biomarker or a therapeutic target for BC.
INTRODUCTION
According to studies from the United States, approximately 74,000 new cases of bladder cancer (BC) are recorded each year that makes it the second most common cancer of genitourinary system after prostate cancer.[1,2,3] In the Indian population, cancer exists with an age-standardized incidence rate of 4.1 in males and 0.4 in females per 100,000 persons.[4,5] BC can be classified into nonmuscle-invasive BC (NMIBC) and muscle-invasive BC (MIBC) on the basis of the degree of invasion of the tumor into deeper muscular layer of the urinary bladder wall. The pathology and clinical behavior of these two groups are different.[5] Although only 20% of patients are diagnosed with MIBC, the majority of deaths are due to its invasion and metastasis. Radical cystectomy is considered as the best procedure for the treatment of MIBC, but about 50% of such patients develop metastases within 2 years after cystectomy and die of the disease.[6] Approximately 75% of patients with NMIBC recur and have 5 year survival rates of 88%–98%.[7] The outcome of NMIBCs is hard to prognosticate based on existing pathological characteristics as they have unpredictable rates of recurrence and progression, so further studies are required to explore the molecular mechanisms and regulations for such unusual oncogenic behavior.
When talking about molecular mechanisms, micro RNA (miRNA)-mediated posttranscriptional gene regulation plays an important role in oncogenesis. The last decade has distinguishably been regarded as the decade of miRNA research to explore possible weapons against various cancers as well as BC.[8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23] However, there is a need for more elaborative research which could make miRNA a potential tool to define BC etiology and to prognosticate the outcome for both MIBC and NMIBC. Apart from their role in diagnosis and prognostication, an area of research into the development of anti-miRNAs could further be explored to develop a therapeutic molecule for the treatment of BC. This review is aimed at comprehensive analysis on progress in miRNA research in BC.
MATERIALS AND METHODS
We have searched PubMed and Google Scholar databases for publications from 1990 to 2016, related to miRNA biogenesis, its function, and role in the pathogenesis of BC as well as other cancers. Articles were searched using MeSH terms micrornas, micrornas AND neoplasm, and micrornas AND urinary bladder neoplasm. In addition, we have searched for articles explaining the role of miRNA in BC as a diagnostic or therapeutic tool. Out of the 108 publication reviewed, 75 references have been cited in this paper. Rest of the articles was excluded due to irrelevant data. Articles were reviewed and compiled to assess the role of miRNA in various cancers and those in BC as a diagnostic or therapeutic tool.
RESULTS
Database search provided more than 100 publications, in which 75 were found to be relevant for this study. Studies have shown that more than 35 miRNAs were associated with different pathways of cellular dedifferentiation, proliferation, and progression of BC as well as other cancers. The biogenesis and functional significance of miRNA, their role in BC as well as other cancers are described in following headings, highlighting their biomarker, and therapeutic potential.
BIOGENESIS AND FUNCTION OF MICRO RNA
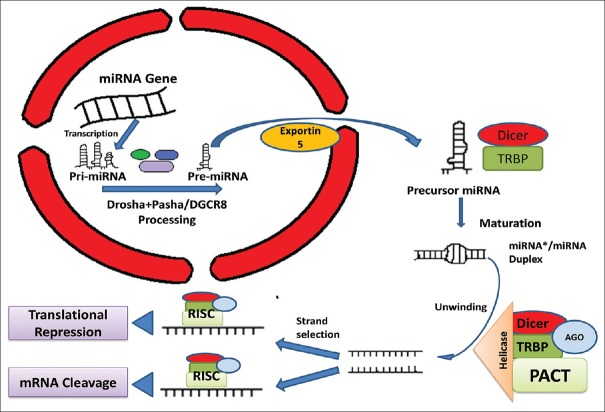
miRNAs are short noncoding single-stranded ribonucleic acids of 20–24 nucleotides in length. Long primary transcripts (pri-miRNAs) transcribed from intronic regions of genomic DNA are modified by RNase type-III enzymes Drosha and Dicer endonuclease to produce pre-miRNAs. Then, mature miRNAs are produced from these pre-miRNAs with the help of a protein complex containing Argonaute and Helicase [Figure 1].[8,9] These small RNAs functions as negative regulators of protein synthesis by pairing with the 3' untranslated region (UTR) of concerned messenger RNAs (mRNAs) with the help of RNA-induced silencing complex [Figure 1].[10] This makes them an important molecule to regulate different metabolic pathways of cellular differentiation and dedifferentiation in all multicellular organisms for example, such as nematodes Caenorhabditis elegans, an arthropod Drosophila melanogaster (invertebrate) or in humans (mammals) and other vertebrates.[8,9,24,25] In cellular physiology, miRNA regulates protein expression by feedback mechanism and from their buffering effects, they provide accuracy to key biological processes. As such, miRNA may control gene expression by decreasing the synthesis of proteins, which are expressed outside their physiologically optimum level.[26,27,28] miRNA targets several mRNAs affecting a multitude of transcripts to control metabolisms associated with cellular growth, division, death and differentiation, influencing numerous cancer-relevant processes such as proliferation, cell cycle checkpoints, apoptosis, dedifferentiation, and metastasis. As we all know that cancer cells are nothing but dedifferentiated cells having persistence division capability. Therefore, before going into the role of miRNAs in pathogenesis of cancer, it is necessary to understand how these tiny molecules promote a cell to lose its identity (de-differentiation) and become an aggressively proliferating cell.
Figure 1.
MiRNA processing and posttranscriptional gene regulation
ROLE OF MICRO RNA IN CELL DIFFERENTIATION
Starting from fertilization and embryonic development to growth, morphogenesis, and stem cell generation of multicellular organisms, there is continuous phenomena of switching OFF and switching ON of several genes, which decides fate of each and every cell, known as cellular differentiation. The cell further stays in its particular morphology and physiology maintaining specific gene pattern.[29]
Regulation of these genetic patterns can be altered by several extracellular signals as well as intracellular posttranscriptional regulation such as RNA interference. The expression of miRNA at an optimum level is important because their loss of function promotes cellular dedifferentiation and oncogenic transformation.[30,31] Stem cells and their differentiation are dependent on protein regulatory mechanism of miRNA.[32] Early studies in worms provided evidence to show that miRNAs serve as regulatory molecules in a particular differentiation pathway.[33] Similarly, in mammalian studies, the role of miR-203 in regulating skin differentiation and miR-143/145 regulating differentiation of smooth muscle cells has also been defined.[34,35] The early overexpression study has also shown that a single miRNA has potential to transform the identity of a particular differentiated cell or tissue into different form.[36] Similarly, miR-304 cluster could induce pluripotency in human and mouse fibroblast driving cells toward transformation.[37,38]
MICRO RNA IN CANCERS
As we know that cancer is nothing but a dedifferentiated mass of cells having uncontrolled capability of proliferation and growth which could be a result of miRNA dysregulation. More than 1400 human miRNAs have been identified till 2010, mostly conserved even among distantly related vertebrates and invertebrates, regulating expression of >60% of all mRNAs.[39,40] The role of miRNAs in cancer pathogenesis is supported by one of the studies, which has shown that most of the genes of miRNA are located in cancer-associated genomic regions or in fragile sites.[11] miRNAs can function as either oncogenes or tumor suppressor genes, likewise oncogenic miRNAs could be overexpressed to target tumor suppressor genes, whereas tumor-suppressor miRNAs could be underexpressed to promote the overexpression of oncogenes.[12] miRNA expression data from in vitro studies on wide spectrum of cancer diseases explained their potential as oncogene, tumor suppressor gene, and epigenetic regulators.[13,14,15,16] Several studies have shown that miRNAs are associated with the regulation of oncogenic pathways influencing the risk of recurrence in BC.[17,18,19,20,21,22,23,41,42] Expression and binding capacity of miR-3646 to rs3242 single nucleotide polymorphism in secreted frizzled-related protein 1 in early age group patients were associated with higher risk for BC.[41] Another recent study has demonstrated a role of miR-127-mediated regulation of PIK3R1 (phosphatidylinositol 3-kinase regulatory subunit alpha) in BC pathways.[17]
One of the interesting features associated with miRNA-mediated gene regulation is that a single miRNA may target several mRNA producing antagonist proteins of the same cellular pathway. Different miRNAs may corepress the same target mRNA also. Hence, there is a need for more extensive studies to resolve these mysteries associated with miRNA-mediated gene regulation. These paradoxes exist due to huge epigenetic diversity of organ-specific tumors, that's why it is difficult to associate the expression of one particular miRNA to oncogenesis.[43,44] Another aspect of miRNA is that their function in oncogenesis may be context dependent. Thus, a particular miRNA may be upregulated in some cancers but downregulated in others, due to different morphological and physiological features of different cell before dedifferentiation. MiR-29 that appears to act as a oncogenic in lung cancer may have tumor-suppressive functions for breast cancer.[45,46] In another example, miR-26a is found to be downregulated in hepatocellular carcinoma, but its overexpression promotes a metastatic phenotype in lung cancer cells.[47,48]
Hence, there is a need to carefully assess particular function of a miRNA to define its role in cancer or other diseases.
BLADDER CANCER BIOLOGY AND MICRO RNA
The unique features of BC are its recurrence and progression. The two broad clinical categories of BC have been defined as NMIBC and MIBC, which have different clinical and pathological characteristics at presentation and have different treatment strategies.[5] One of the probable hypotheses for the existence of these two categories is miRNA-mediated gene regulation like we discussed earlier in cell differentiation.
Expression of different miRNAs and their functional role have been studied in BC along with their clinical features such as stages and grades.[18] A recent study in 2013 reported that expression of miR-23b was low in BC tissue in comparison to the normal one, which could function as tumor suppressor by regulating Zinc finger E-box 1.[19] Similarly, fibroblast growth factor receptor signaling has a major role in differentiation of cell and their pathways and have also been found to be dysregulated in BC. MiR-129, miR-145, miR101, miR99a, miR-100, miR-7, miR-203, and miR205 were found to orchestrate expression of different proteins in BC.[18] The role of miRNA-7 as tumor suppressor was also demonstrated by showing a SNP (1010A/G) at 3' UTR (target of miR-7) of Homeo Box B5 gene which may act as an oncogene.[20] Another study explained miR-182-5p to downregulate RECK and Smad4 showing its oncogenic nature.[21]
miRNA expression profile in BC tissues and their normal counterpart has been compared and correlated with different stages to prognosticate the outcome.[22,42] Expression of miR-200 family has been shown as a marker of progression in invasive BC.[23] In one of the recent studies from our laboratory, we have shown that there was differential expression of miR-129, miR-200a, and miR-205 between normal looking mucosa of a BC patient and that of a healthy person.[49]
Micro RNA studies in bladder cancer
The studies of miRNAs in BC can be broadly classified into two groups, i.e. mechanistic studies (in vitro and in vivo studies of forced miRNA expression either in cell lines or in animal models) and observational studies (miRNA expression in BC patients and their association with disease parameters to elucidate their biomarker and therapeutic potential) [Table 1].
Table 1.
Micro RNA studies in bladder cancer

Mechanistic studies
To elucidate the role of micro RNA in bladder cancer pathogenesis
Broadly, miRNA could act in two ways, i.e., either to downregulate a tumor suppressor gene or to downregulate an oncogene. To further elucidate the mechanism of miRNA-mediated gene regulation, cell lines of the urinary bladder epithelium have been extensively used for in vitro experiments. miRNA expression profile of T24 BC cell line provides evidence that miR-127 may have a tumor suppressive function.[50] The miR-200 family maintains epithelial phenotype and its transfection in BC cell lines induces the expression of epithelial proteins and suppression of mesenchymal proteins.[51] Similarly, highly expressed Ras oncogene in BC was found to be deregulated by miR-143 in EJ and T24 human BC cell lines.[52] Apoptotic cell death was also shown on exogenous expression of miR-145.[19,53]
Observational studies
Studies showing association of micro RNA profile with bladder cancer profile
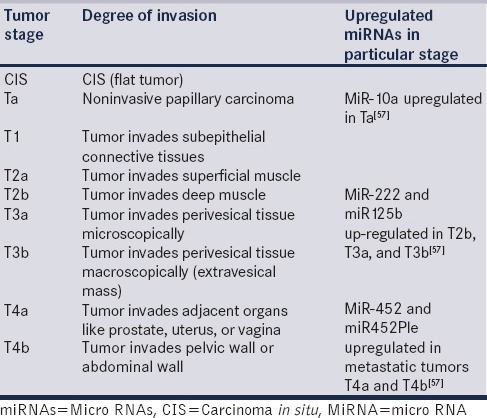
Aggressiveness of the different phenotypes of BC can be classified into different stages and grades [Table 2]. It is a general hypothesis that being a regulatory molecule, expression of miRNAs should vary in different stages and grades. The expression of miR-159-3p, which was shown to be downregulated in BC tissue as compared with adjacent normal tissues was found to be negatively correlated with grades.[54] It is not always necessary that expression of miRNA would correlate with stage and grade of the tumor because during the dedifferentiation process, there could be switching “ON” and “OFF” of many genes, which give unique morphology and physiology to tumors to survive in the vicinity. Some of the miRNAs may be present/absent or over/under expressed at specific stage and grade, like miR-145 is under-expressed in low-grade, noninvasive, and high-grade invasive tumors.[55] Alteration in miRNA expression could be a characteristic feature of high and low grade BC.[18] miR-182, miR-183, miR-10a, miR-203, and miR-224 were all found to be significantly upregulated whereas miR-1, miR-143, miR-145, miR-133a, miR-133b, and miR-125b were found to be significantly downregulated in BC.[42,56] miR-222 and miR125b were found to be upregulated in MIBC and miR-10a in NMIBC [Table 2]. Further, miR-452 and miR-452* were found to be upregulated in metastatic tumors of higher stage and grade.[57]
Table 2.
Stages of bladder tumor and some of the up-regulated micro RNAs
Besides stages and grades for tumor aggressiveness, NMIBC is known for its multifocality (tumors arising from different locations in the bladder) and higher recurrence rate. A normal looking mucosa may show molecular changes preceding phenotypic changes in the form of a visible tumor. In a study done at our center, we found varied expression of miR-129, miR-200a, and miR-205 between normal looking mucosa of a BC patient and that of a healthy person.[49] Therefore, normal looking mucosa of patients with bladder tumor may not be genetically normal. Genetic changes precede phenotypic changes, which could be the reason for recurrence of tumor in different zones of bladder epithelium. There is a need for elaborative studies, which could profile miRNA expression in recurrent tumor for better prognosis of the outcome. In this regard, one of the recent studies in 2013 classified two groups of NMIBC, i.e., one in which tumor progressed to muscle invasive tumors and the other in which tumor did not progress. They have shown that the expression of miR-29c was downregulated in tumors that progressed to muscle invasive. Hence, miR-29c could be used to identify patients with tumors of T1 stage and high grades for potential risk of muscle invasion.[58] Expression of miR-3646 has also been shown to be associated with risk and recurrence of BC.[41]
Metastasis is the ultimate outcome of MIBC. Cancer cells in mesenchyme or circulatory system are responsible for the metastasis of a localized cancer. Cellular metabolic pathways associated with proliferation and epithelial-mesenchymal transition (EMT) is generally hyperactive during the progression of a cancer to metastatic stage. Different miRNA regulatory mechanisms for EMT and metastasis in BC have been studied. miR-221 has been demonstrated to facilitate transforming growth factor (TGF-β1) mediated EMT in BC.[59] ROCK1 is a protein, which promotes EMT and metastasis and found to be deregulated by miR-124-3p and miR-1280 explaining their potential to suppress metastasis of BC.[60,61] MiR-590-3p and miR-574-3p have also been recognized as metastatic suppressors.[54,62] miRNAs, instead of suppressor of metastasis, they could also induce metastasis, such as MiR-182-5p has been demonstrated to be promoting cell viability, migration, invasion, and inhibition of apoptosis on its overexpression.[21]
Studies showing association of micro RNA profile with survival
As described earlier miRNAs are responsible for the regulation of tumor progression and metastasis and thus could be directly correlated with the survival of the patient. Overexpression of miR-23b has been found to be positively correlated with higher overall survival while overexpression of miR-182-5p has been shown to be associated with reduced survival.[19,21] Downregulation of miR-100 and upregulation of miR-222 have also been shown to be independently associated with shorter progression-free survival and overall survival of BC patients.[63,64]
Studies showing potential of micro RNA as biomarker
Cystoscopy despite being invasive procedure is considered as the gold standard for detecting recurrence in NMIBC. Keeping the fact in mind that all cancers show miRNA dysregulation, there are studies going on in search of specific miRNA as molecular biomarker for prognostication of BC.[13,14,15,16,22,65] In vitro studies on BC cell lines and mouse model-based studies have shown the potential of miRNA as biomarker.[57,66] MiR-23b, miR-182-5p, miR-10a, miR-125b, and miR-222 have been identified as biomarkers on the basis of their expression level in malignant and nonmalignant tissues.[19,21,57] Expression of miR-10a, miR-125b, and miR-222 have been found to be specific for muscle-invasive tumors.[57] Hence, depending on the expression of such miRNAs, one could plan the protocol of surveillance by altering the number of cystoscopic examination and thereby reducing the frequent visit to the clinic for an invasive test like cystoscopy to detect recurrence. Urine is an ideal body fluid to detect a biomarker which could tell us about the recurrence of BC. There is a possibility that miRNAs might be present in patient's urine as it remains in contact with tumor for a significant period. miR-214, miR-99a, and miR-125b have been shown to be potential urinary biomarkers for screening as well as predicting recurrences in NMIBC.[67,68] MiR-497 and miR-663b were found to be circulating biomarkers in clinical detection of BC.[69] miRNA-141 and miRNA-200b having roles in the invasive ability and EMT phenotype could also help biomarker to identify patients undergoing cystectomy who are likely to have lymph node metastasis.[70]
Studies showing therapeutic potential of micro RNA
Chemotherapy and drug-based therapy are being studied by targeting protein molecules either, ligand, receptor or intermediate metabolite of signaling pathways, responsible for cellular proliferation, migration, and invasion. miRNAs could act as a repressor for protein molecule functioning or activator of different pathways, according to cellular metabolic demands. Thus, considering these natural regulators as diagnostic as well as therapeutic molecules, they could be part of a new area of research.[57,66,71,72] The two possible approaches by which researcher could investigate therapeutic potential of these molecules would be, either using a miRNA, which targets an oncogene or using an anti-miRNA, which targets an overexpressed miRNA against tumor suppressor gene. A protein survivin, which was upregulated in high-grade BC, could be shown to be downregulated in miR-1826 transfected BC cell lines.[21,73] In the same study, three oncogenes, i.e., beta-catenin, mitogen-activated protein kinase 1, and vascular endothelial growth factor C were shown as targets of miR-1826, which was confirmed through Western blotting technique.[21] Limitations to miRNA-based cancer therapy such as unstable nature of miRNA, their rapid plasma clearance and poor uptake into the tumor cells are challenging.[74] There have been efforts to overcome these challenges for miRNAs to reach to the target cells in vivo. Lipid-based vesicles such as stable nucleic acid-lipid particles having stability in serum and good transfection efficiency could be potential delivery system for tumor suppressive miRNAs.[75]
CONCLUSIONS
BC has unique characteristics of presentation, wherein NMIBC are notorious for recurrence and progression and MIBC have not yet shown a good outcome in general. Existing clinicopathological characteristics of BC are not sufficient to meet the desired goals of prediction and prognostication. miRNA is a new ray of hope in finding out a new marker for surveillance in NMIBC after initial endoscopic resection of the tumor with the aim of replacing regular cystoscopy to look for recurrence. At the same time, miRNAs could serve as target for therapeutic purpose to get survival advantage in MIBC. With these two clinical goals, this review article would strengthen the potential application of miRNA research in understanding the bladder tumor biology and would give a new direction in translational research.
Footnotes
Financial support and sponsorship: Author Nilay Mitash was supported by a fellowship grant from Council of Scientific and Industrial Research (CSIR), New Delhi, India.
Conflicts of interest: There are no conflicts of interest.
REFERENCES
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Oosterlinck W, Lobel B, Jakse G, Malmström PU, Stöckle M, Sternberg C. European Association of Urology (EAU) Working Group on Oncological Urology. Guidelines on bladder cancer. Eur Urol. 2002;41:105–12. doi: 10.1016/s0302-2838(01)00026-4. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–22. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manoharan N, Tyagi BB, Raina V. Cancer incidences in urban Delhi-2001-05. Asian Pac J Cancer Prev. 2009;10:799–806. [PubMed] [Google Scholar]
- 5.Manoharan N, Tyagi BB, Raina V. Cancer incidences in rural Delhi-2004-05. Asian Pac J Cancer Prev. 2010;11:73–7. [PubMed] [Google Scholar]
- 6.Babjuk M, Oosterlinck W, Sylvester R, Kaasinen E, Böhle A, Palou-Redorta J. European Association of Urology (EAU). EAU guidelines on non-muscle-invasive urothelial carcinoma of the bladder. Eur Urol. 2008;54:303–14. doi: 10.1016/j.eururo.2008.04.051. [DOI] [PubMed] [Google Scholar]
- 7.Proctor I, Stoeber K, Williams GH. Biomarkers in bladder cancer. Histopathology. 2010;57:1–13. doi: 10.1111/j.1365-2559.2010.03592.x. [DOI] [PubMed] [Google Scholar]
- 8.Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, et al. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- 9.Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, et al. MicroRNA-responsive 'sensor' transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–83. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- 10.Seitz H, Youngson N, Lin SP, Dalbert S, Paulsen M, Bachellerie JP, et al. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–2. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- 11.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 13.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munker R, Calin GA. MicroRNA profiling in cancer. Clin Sci (Lond) 2011;121:141–58. doi: 10.1042/CS20110005. [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 17.Xu Y, Luo S, Liu Y, Li J, Lu Y, Jia Z, et al. Integrated gene network analysis and text mining revealing PIK3R1 regulated by miR-127 in human bladder cancer. Eur J Med Res. 2013;18:29. doi: 10.1186/2047-783X-18-29. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Catto JW, Miah S, Owen HC, Bryant H, Myers K, Dudziec E, et al. Distinct microRNA alterations characterize high-and low-grade bladder cancer. Cancer Res. 2009;69:8472–81. doi: 10.1158/0008-5472.CAN-09-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Majid S, Dar AA, Saini S, Deng G, Chang I, Greene K, et al. MicroRNA-23b functions as a tumor suppressor by regulating Zeb1 in bladder cancer. PLoS One. 2013;8:e67686. doi: 10.1371/journal.pone.0067686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo J, Cai Q, Wang W, Huang H, Zeng H, He W, et al. A microRNA-7 binding site polymorphism in HOXB5 leads to differential gene expression in bladder cancer. PLoS One. 2012;7:e40127. doi: 10.1371/journal.pone.0040127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirata H, Ueno K, Shahryari V, Tanaka Y, Tabatabai ZL, Hinoda Y, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS One. 2012;7:e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, et al. Genomic profiling of microRNAs in bladder cancer: Mir-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–60. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 23.Wiklund ED, Bramsen JB, Hulf T, Dyrskjøt L, Ramanathan R, Hansen TB, et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int J Cancer. 2011;128:1327–34. doi: 10.1002/ijc.25461. [DOI] [PubMed] [Google Scholar]
- 24.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 25.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 26.Herranz H, Cohen SM. MicroRNAs and gene regulatory networks: Managing the impact of noise in biological systems. Genes Dev. 2010;24:1339–44. doi: 10.1101/gad.1937010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 28.Levine E, McHale P, Levine H. Small regulatory RNAs may sharpen spatial expression patterns. PLoS Comput Biol. 2007;3:e233. doi: 10.1371/journal.pcbi.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ben-Tabou de-Leon S, Davidson EH. Gene regulation: Gene control network in development. Annu Rev Biophys Biomol Struct. 2007;36:191. doi: 10.1146/annurev.biophys.35.040405.102002. [DOI] [PubMed] [Google Scholar]
- 30.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–7. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 31.Peter ME. Let-7 and miR-200 microRNAs: Guardians against pluripotency and cancer progression. Cell Cycle. 2009;8:843–52. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich EM, Dimmeler S. MicroRNAs and stem cells: Control of pluripotency, reprogramming, and lineage commitment. Circ Res. 2012;110:1014–22. doi: 10.1161/CIRCRESAHA.111.243394. [DOI] [PubMed] [Google Scholar]
- 33.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 34.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing 'stemness'. Nature. 2008;452:225–9. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–10. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 37.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sotiropoulou G, Pampalakis G, Lianidou E, Mourelatos Z. Emerging roles of microRNAs as molecular switches in the integrated circuit of the cancer cell. RNA. 2009;15:1443–61. doi: 10.1261/rna.1534709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr Protoc Bioinformatics. 2010;12:9. doi: 10.1002/0471250953.bi1209s29. [DOI] [PubMed] [Google Scholar]
- 40.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rogler A, Hoja S, Socher E, Nolte E, Wach S, Wieland W, et al. Role of two single nucleotide polymorphisms in secreted frizzled related protein 1 and bladder cancer risk. Int J Clin Exp Pathol. 2013;6:1984–98. [PMC free article] [PubMed] [Google Scholar]
- 42.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–9. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 43.Alvarez-Saavedra E, Horvitz HR. Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol. 2010;20:367–73. doi: 10.1016/j.cub.2009.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsang JS, Ebert MS, van Oudenaarden A. Genome-wide dissection of microRNA functions and cotargeting networks using gene set signatures. Mol Cell. 2010;38:140–53. doi: 10.1016/j.molcel.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–5. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kota J, Chivukula RR, O'Donnell KA, Wentzel EA, Montgomery CL, Hwang HW, et al. Therapeutic microRNA delivery suppresses tumorigenesis in a murine liver cancer model. Cell. 2009;137:1005–17. doi: 10.1016/j.cell.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu B, Wu X, Liu B, Wang C, Liu Y, Zhou Q, et al. MiR-26a enhances metastasis potential of lung cancer cells via AKT pathway by targeting PTEN. Biochim Biophys Acta. 2012;1822:1692–704. doi: 10.1016/j.bbadis.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 49.Mitash N, Agnihotri S, Mittal B, Tiwari S, Mandhani A. Molecular cystoscopy: Micro-RNAs could be a marker for identifying genotypic changes for transitional cell carcinoma of the urinary bladder. Indian J Urol. 2016;32:149–53. doi: 10.4103/0970-1591.174775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006;5:2220–2. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 51.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin T, Dong W, Huang J, Pan Q, Fan X, Zhang C, et al. MicroRNA-143 as a tumor suppressor for bladder cancer. J Urol. 2009;181:1372–80. doi: 10.1016/j.juro.2008.10.149. [DOI] [PubMed] [Google Scholar]
- 53.Noguchi S, Yamada N, Kumazaki M, Yasui Y, Iwasaki J, Naito S, et al. socs7, a target gene of microRNA-145, regulates interferon-ß induction through STAT3 nuclear translocation in bladder cancer cells. Cell Death Dis. 2013;4:e482. doi: 10.1038/cddis.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mo M, Peng F, Wang L, Peng L, Lan G, Yu S. Roles of mitochondrial transcription factor A and microRNA-590-3p in the development of bladder cancer. Oncol Lett. 2013;6:617–623. doi: 10.3892/ol.2013.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dip N, Reis ST, Srougi M, Dall'Oglio MF, Leite KR. Expression profile of microrna-145 in urothelial bladder cancer. Int Braz J Urol. 2013;39:95–101. doi: 10.1590/S1677-5538.IBJU.2013.01.12. [DOI] [PubMed] [Google Scholar]
- 56.Ichimi T, Enokida H, Okuno Y, Kunimoto R, Chiyomaru T, Kawamoto K, et al. Identification of novel microRNA targets based on microRNA signatures in bladder cancer. Int J Cancer. 2009;125:345–52. doi: 10.1002/ijc.24390. [DOI] [PubMed] [Google Scholar]
- 57.Veerla S, Lindgren D, Kvist A, Frigyesi A, Staaf J, Persson H, et al. MiRNA expression in urothelial carcinomas: Important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–42. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 58.Rosenberg E, Baniel J, Spector Y, Faerman A, Meiri E, Aharonov R, et al. Predicting progression of bladder urothelial carcinoma using microRNA expression. BJU Int. 2013;112:1027–34. doi: 10.1111/j.1464-410X.2012.11748.x. [DOI] [PubMed] [Google Scholar]
- 59.Liu J, Cao J, Zhao X. miR-221 facilitates the TGFbeta1-induced epithelial-mesenchymal transition in human bladder cancer cells by targeting STMN1. BMC Urol. 2015;15:36. doi: 10.1186/s12894-015-0028-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Li S, Lin Y, Chen H, Hu Z, Mao Y, et al. MicroRNA-124-3p inhibits cell migration and invasion in bladder cancer cells by targeting ROCK1. J Transl Med. 2013;11:276. doi: 10.1186/1479-5876-11-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang S, Xue S, Dai Y, Yang J, Chen Z, Fang X, et al. Reduced expression of microRNA-100 confers unfavorable prognosis in patients with bladder cancer. Diagn Pathol. 2012;7:159. doi: 10.1186/1746-1596-7-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tatarano S, Chiyomaru T, Kawakami K, Enokida H, Yoshino H, Hidaka H, et al. Novel oncogenic function of mesoderm development candidate 1 and its regulation by MiR-574-3p in bladder cancer cell lines. Int J Oncol. 2012;40:951–9. doi: 10.3892/ijo.2011.1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YK, Kim WJ. Epigenetic markers as promising prognosticators for bladder cancer. Int J Urol. 2009;16:17–22. doi: 10.1111/j.1442-2042.2008.02143.x. [DOI] [PubMed] [Google Scholar]
- 64.Zhang DQ, Zhou CK, Jiang XW, Chen J, Shi BK. Increased expression of miR-222 is associated with poor prognosis in bladder cancer. World J Surg Oncol. 2014;12:241. doi: 10.1186/1477-7819-12-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: Progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849–64. doi: 10.1038/nrc3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim SM, Kang HW, Kim WT, Kim YJ, Yun SJ, Lee SC, et al. Cell-free microRNA-214 from urine as a biomarker for non-muscle-invasive bladder cancer. Korean J Urol. 2013;54:791–6. doi: 10.4111/kju.2013.54.11.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang DZ, Lau KM, Chan ES, Wang G, Szeto CC, Wong K, et al. Cell-free urinary microRNA-99a and microRNA-125b are diagnostic markers for the non-invasive screening of bladder cancer. PLoS One. 2014;9:e100793. doi: 10.1371/journal.pone.0100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Du M, Shi D, Yuan L, Li P, Chu H, Qin C, et al. Circulating miR-497 and miR-663b in plasma are potential novel biomarkers for bladder cancer. Sci Rep. 2015;5:10437. doi: 10.1038/srep10437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liu W, Qi L, Lv H, Zu X, Chen M, Wang J, et al. MiRNA-141 and miRNA-200b are closely related to invasive ability and considered as decision-making biomarkers for the extent of PLND during cystectomy. BMC Cancer. 2015;4(15):92. doi: 10.1186/s12885-015-1110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tili E, Michaille JJ, Gandhi V, Plunkett W, Sampath D, Calin GA. miRNAs and their potential for use against cancer and other diseases. Future Oncol. 2007;3:521–37. doi: 10.2217/14796694.3.5.521. [DOI] [PubMed] [Google Scholar]
- 72.Trang P, Weidhaas JB, Slack FJ. MicroRNAs as potential cancer therapeutics. Oncogene. 2008;27(Suppl 2):S52–7. doi: 10.1038/onc.2009.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horstmann M, Bontrup H, Hennenlotter J, Taeger D, Weber A, Pesch B, et al. Clinical experience with survivin as a biomarker for urothelial bladder cancer. World J Urol. 2010;28:399–404. doi: 10.1007/s00345-010-0538-2. [DOI] [PubMed] [Google Scholar]
- 74.Bravo V, Rosero S, Ricordi C, Pastori RL. Instability of miRNA and cDNAs derivatives in RNA preparations. Biochem Biophys Res Commun. 2007;353:1052–5. doi: 10.1016/j.bbrc.2006.12.135. [DOI] [PubMed] [Google Scholar]
- 75.Semple SC, Klimuk SK, Harasym TO, Dos Santos N, Ansell SM, Wong KF, et al. Efficient encapsulation of antisense oligonucleotides in lipid vesicles using ionizable aminolipids: Formation of novel small multilamellar vesicle structures. Biochim Biophys Acta. 2001;1510:152–66. doi: 10.1016/s0005-2736(00)00343-6. [DOI] [PubMed] [Google Scholar]