Figure 1. G6PD variant classes are distributed differently across the G6PD structure.
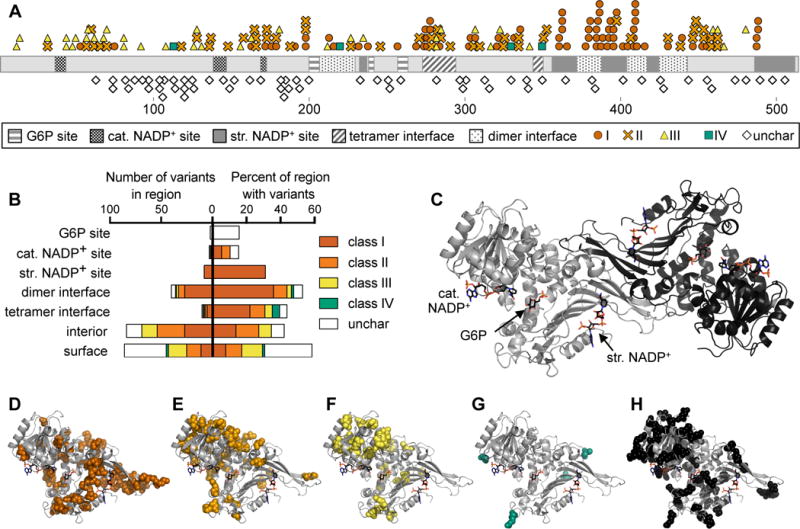
A) Linear representation of G6PD showing location of variants and structural regions. Structural regions shown are the G6P, catalytic (cat.) NADP+, and structural (str.) NADP+ binding sites, and dimer and tetramer interfaces. Variants shown are class I–IV and uncharacterized (unchar) variants from the ExAC database.
B) Quantification of the number of variants in each structural region (left) and the percent of amino acids in each region for which a variant has been identified (right).
C) Crystal structure of dimeric G6PD, assembled from PDB IDs 2BH9 and 2BHL.
D–H) Variant locations are shown in spheres on the monomeric structure of G6PD: (D) class I, (E) class II, (F) class III, (G) class IV, (H) uncharacterized variants from the ExAC database.
