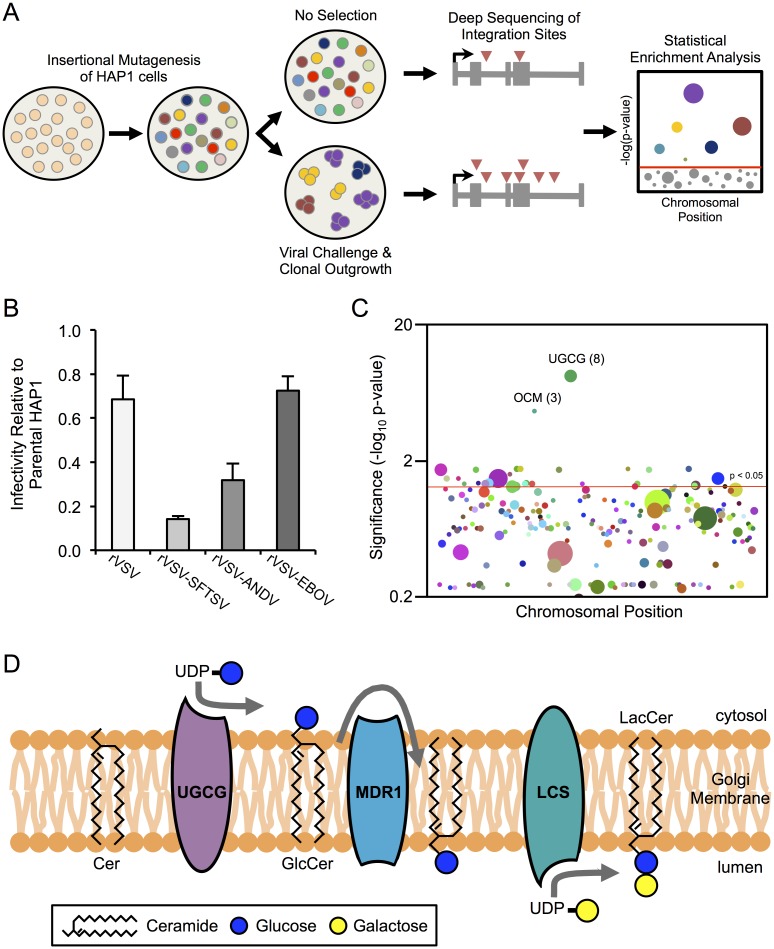
Fig 1. Haploid genetic screen identifies SFTSV entry factors.
(A) Overview of haploid genetic screen. Details provided in Materials and Methods. (B) Parental (non-mutagenized) HAP1 cells and mutagenized HAP1 cells surviving rVSV-SFTSV selection (HAP1-SFTSVR) were subject to a single-cycle infection (8–10hr) with a panel of recombinant vesicular stomatitis viruses (rVSVs) encoding various viral glycoproteins. Infection in HAP1-SFTSVR cells is normalized to infection in the parental HAP1 cells. SFTSV = severe fever with thrombocytopenia syndrome virus, ANDV = Andes virus, EBOV = Ebola virus. (C) Statistical Enrichment Analysis was carried out using the Chi-Square Exact Test with false discovery rate correction. Each dot represents a unique gene identified from integration site mapping in the HAP1-SFTSVR population. The size of the dot, and number in parenthesis when provided, reflects the number of unique integration sites found within that particular gene. For clarity, only genes with 3 or more unique integration sites are plotted. (D) Schematic of glucosylceramide synthesis. GlcCer is synthesized by the membrane-bound UGCG on the cytosolic face of the cis-Golgi. The polar head of GlcCer is then flipped to the lumenal side by the transporter MDR1, where it can be further modified by lactosylceramide synthase (LCS) to form lactosylceramide, the core building block of most cellular glycosphingolipids.