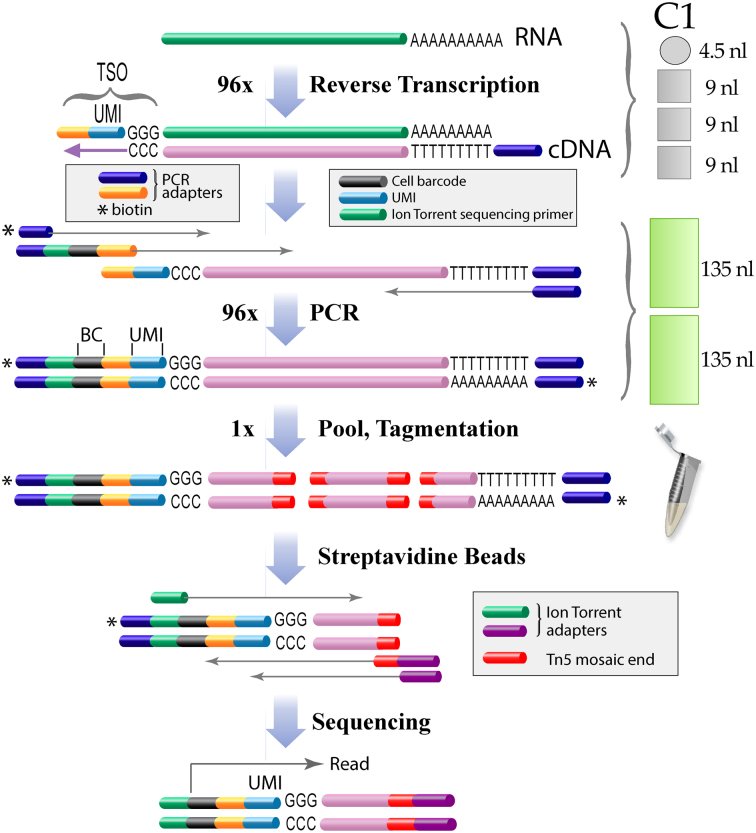
Figure 1.
On chip barcoding workflow. After cell lysis in 4.5 nl poly-adenylated RNA is reverse-transcribed in 31.5 nl with an anchored oligodT primer. A PCR primer sequence and unique molecular identifiers (UMIs) are added to the 3΄ end of the cDNA via reverse transcriptase template switching. The cDNA is subsequently amplified and cell index sequences (barcode) as well as terminal biotins are introduced by PCR in the microfluidic device. The barcoded cDNAs are pooled, fragmented by tagmentation with Tn5 transposase and the biotinylated terminal fragments are isolated on streptavidin beads. 5΄ terminal fragments are selectively amplified and additional sequences required for Ion Torrent sequencers are introduced by PCR. For a detailed protocol see Supplementary Data and for Illumina sequencers see Supplementary Data.