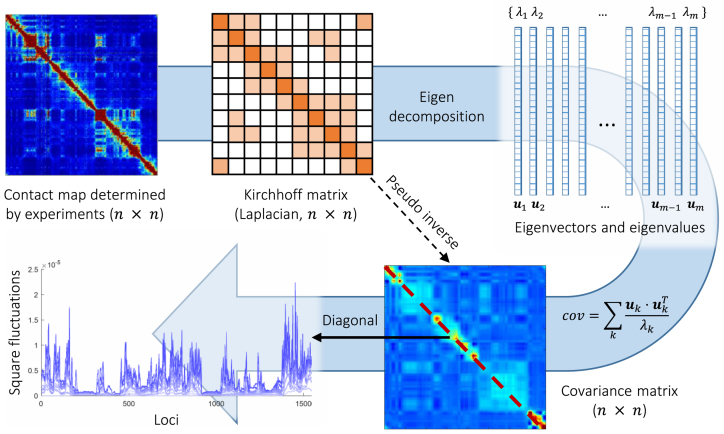
Figure 1.
Schematic description of GNM methodology applied to Hi-C data. The inter-loci contact data represented by the Hi-C map (upper left, for 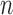 genomic bins (loci)) is used to construct the GNM Kirchhoff matrix,
genomic bins (loci)) is used to construct the GNM Kirchhoff matrix, 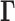 (top, middle). Eigenvalue decomposition of
(top, middle). Eigenvalue decomposition of 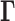 yields a series of eigenmodes which are used for computing the covariance matrix (lower, right), the diagonal elements of which reflect the mobility profile of the loci (bottom, left), and the off-diagonal elements provide information on locus-locus spatial cross-correlations.
yields a series of eigenmodes which are used for computing the covariance matrix (lower, right), the diagonal elements of which reflect the mobility profile of the loci (bottom, left), and the off-diagonal elements provide information on locus-locus spatial cross-correlations. 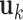 ,
, 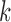 th eigenvector;
th eigenvector; 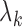 ,
, 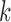 th eigenvalue;
th eigenvalue; 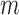 , number of nonzero modes, starting from the lowest-frequency mode, included in the GNM analysis (
, number of nonzero modes, starting from the lowest-frequency mode, included in the GNM analysis (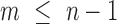 ). In the present application to the chromosomes,
). In the present application to the chromosomes, 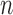 varies in the range
varies in the range 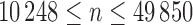 , the lower and upper limits corresponding respectively to chromosomes 22 and 1.
, the lower and upper limits corresponding respectively to chromosomes 22 and 1.