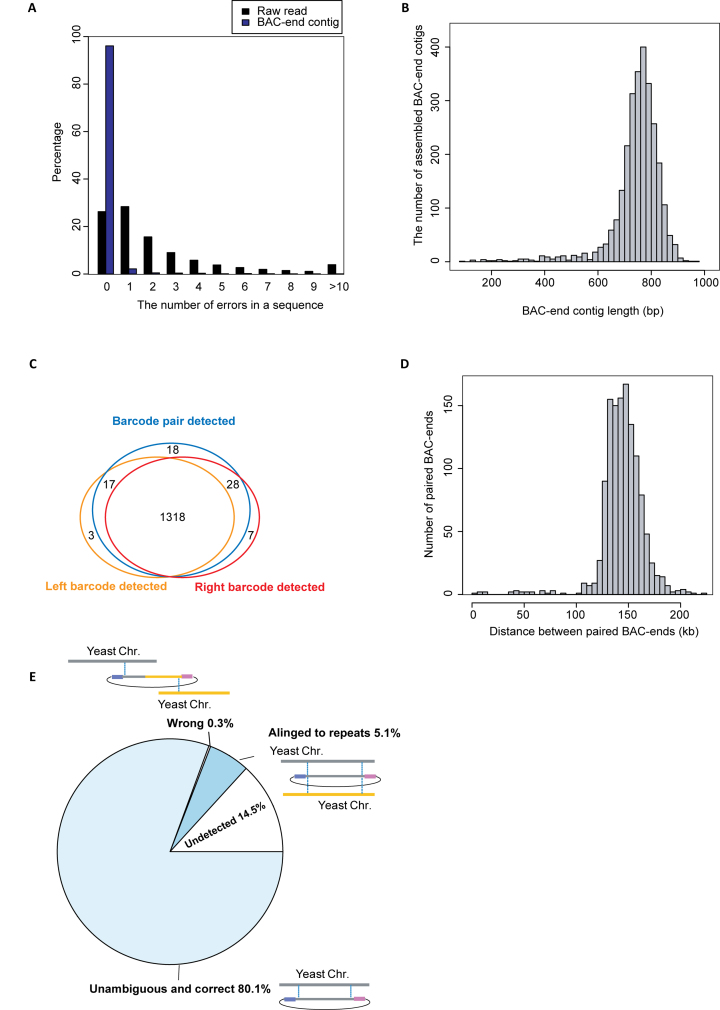
Figure 3.
Characterization of BAC-ends from the yeast BAC library. (A) The distribution of sequencing errors in filtered raw reads and BAC-end contigs, respectively. BAC-end contigs were derived from local assembly using 0.45 M reads encompassing 1373 barcodes for the left ends, and 0.45 M reads encompassing 1387 barcodes for the right ends. (B) The length distribution of the BAC-end contigs. (C) Barcode detection coverage. Left and right barcodes were sequenced with their linked BAC ends using our BAC-end sequencing protocol (Figure 2A). Barcode pairs were detected using our barcode pair sequencing protocol (Figure 2B). (D) The length distribution of genomic fragments spanned by unambiguous and correct paired BAC-end sequences. (E) Categorization of yeast BAC paired-ends. A BAC clone was considered undetected if either barcode was missing after BAC-end sequencing. A BAC clone was considered unambiguous and correct if its ends were aligned to two unique genomic loci spanning <300 kb apart in convergent orientation. An incorrect BAC clone is one whose paired-ends aligned to unique genome loci but on different chromosomes, or not in the convergent orientation on same chromosome, or more than 300 kb apart on same chromosome.