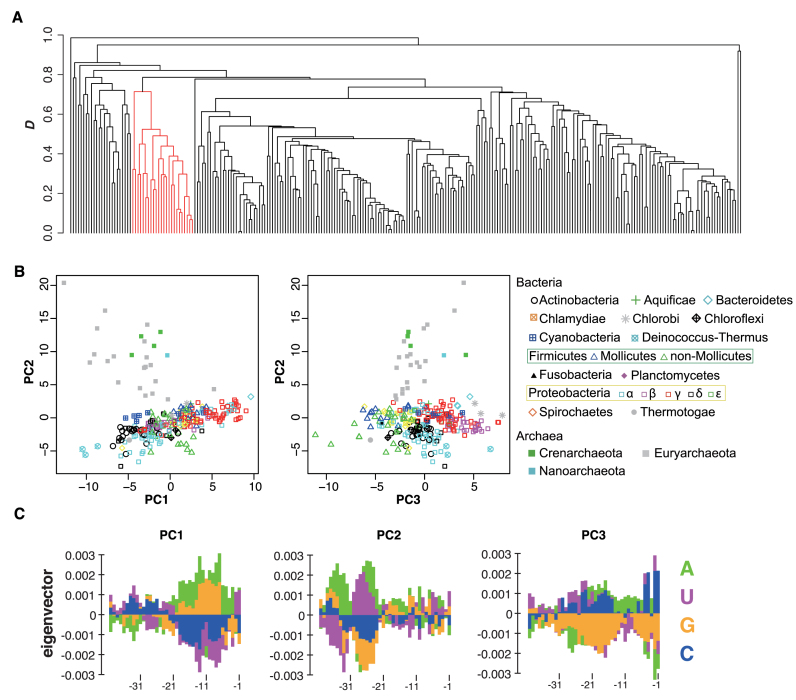
Figure 1.
Cluster and principal component analyses of the upstream regions of non-Shine-Dalgarno (SD) genes for each species. (A) Cluster dendrogram showing similarities in the patterns of gn values upstream of the initiation codon from –40 to –1 among 260 species according to the score D. Branches in red represent a cluster containing all archaeal species. (B) Scatter plots of principal components (PCs) 1 and 2, and 2 and 3. The symbols indicate each taxonomic group of species. Proteobacteria were subdivided into five classes (α, β, γ, δ, and ε), because of the large number of species included in this phylum (138 species). Moreover, Firmicutes were subdivided into two classes, Mollicutes and others, because the fractions of SD genes in Mollicutes including Mycoplasma spp., are significantly lower than those in other Firmicutes (6). (C) The eigenvectors of PC1, 2, and 3 from left to right. The color scheme for each nucleotide is shown in the bottom right. See Supplementary Figure S1 for details of G-statistics analysis.