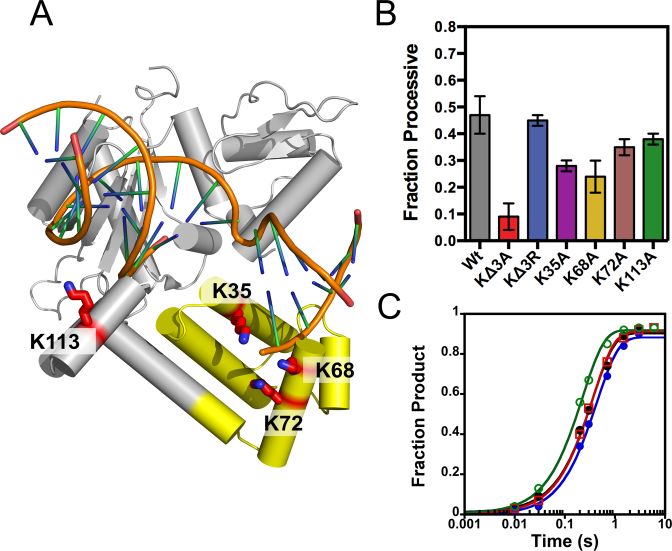
Figure 6.
Pol β uses the positively charged 8-kDa lyase domain for processive searching. Crystal structure of Pol β bound to a 1-nt gapped DNA (PDB 3ISB). (A) The lysine residues mutated to alanine are shown as red sticks. Lysines 35, 68 and 72 reside in the lyase domain (yellow) and K113 within the 31-kDa domain (gray). (B) The fraction processive at 100 mM ionic strength was measured using the P20 substrate with each mutant Pol β under standard reaction conditions. The mean and standard deviation is shown for three independent experiments. (C) Single-turnover analysis for Pol β catalyzed nucleotide insertion measured with indicated mutant enzymes at 100 mM ionic strength. The Wt data is shown as black circles, KΔ3A as red squares, KΔ3R as blue circles, and K113A as green open circles. The nucleotide insertion rate constant (kpol) is comparable among the variant enzymes: Wt (2.8 ± 0.2 s−1), KΔ3A (2.3 ± 0.2 s−1), KΔ3R (2.8 ± 0.2 s−1) and K113A (4.4 ± 0.2 s−1).