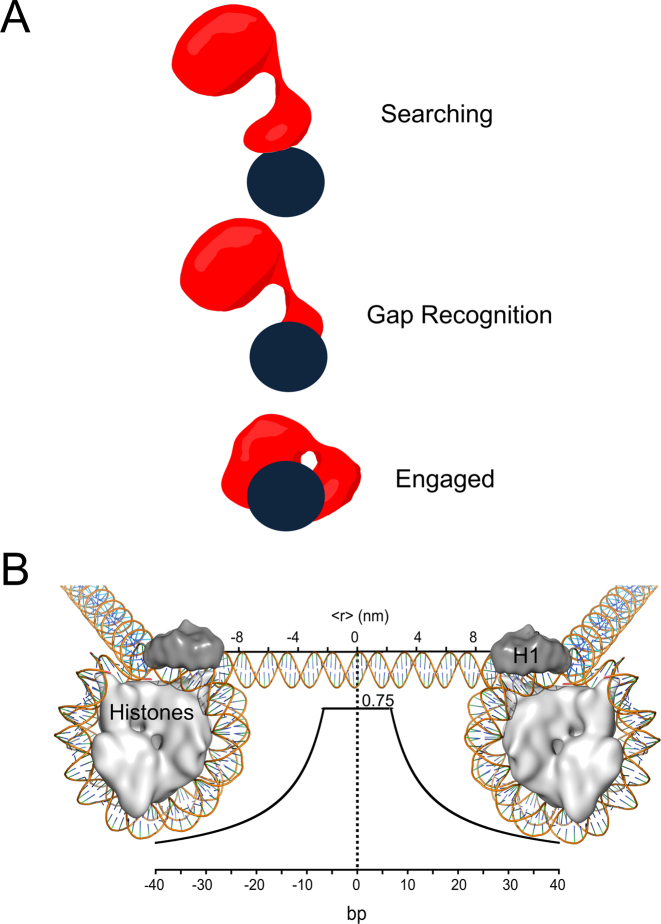
Figure 8.
Models of Pol β searching and gap recognition. (A) Proposed modes of Pol β substrate search and recognition. The dark blue circle represents DNA and is positioned such that the viewpoint is looking down DNA. Pol β is shown as a red cartoon. In the searching mode, Pol β is proposed to mainly use its lyase domain to scan DNA in a hopping mechanism. Once a gap is encountered, the lyase domain makes specific interactions with gapped DNA, allowing for the 31-kDa domain to engage. (B) The probability of Pol β locating and catalyzing nucleotide insertion within a single DNA binding encounter near predicted physiological ionic strength. The curve represents the fit to the hopping equation shown in Figure 5. The y-axis represents the probability, or fraction processive (0-1), of Pol β successfully locating and catalyzing nucleotide insertion. The bottom x-axis represents the number of base pairs and the top axis is the corresponding distance (nm). An approximant DNA linker length of 56 bps is shown for reference. This model can be most easily interpreted by using the origin as the site of DNA damage (a 1-nt gap). A horizontal line ∼6 bp down and up-stream from the origin at a probability of 0.75 indicates that when Pol β binds within 6 bp of damage it has a 0.75 chance of catalyzing nucleotide insertion (Supplementary Figure S4). If Pol β binds ∼10 bp down or up-stream from a 1-nt gap it has ∼50% chance of locating and catalyzing nucleotide insertion. Of the Pol β molecules that are processive, 50% will travel a distance of ∼12 bp down or up-stream, resulting in a mean search footprint of ∼24 bp.