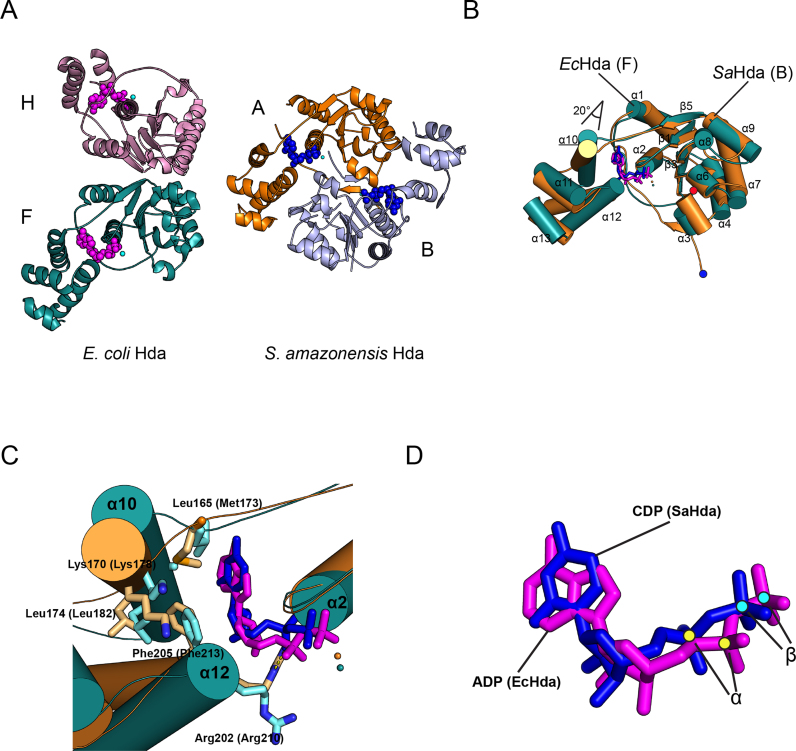
Figure 5.
Comparison of the structures of the Escherichia coli Hda dimer with the Shewanella amazonensis Hda dimer. (A) The E. coli Hda dimer (chain F, teal; chain H, pink) in the Hda–β clamp complex is shown. Nucleotides are shown as pink spheres (left). The S. amazonensis Hda dimer (chain A, light blue; chain B, orange) is also shown. Nucleotides are shown in blue. The F chain of EcHda and the A chain of SaHda are displayed in the same orientation. (B) The superimposed structures of EcHda (chain F, teal) of the Hda–β clamp complex and SaHda (3BOS; chain A, orange) together with secondary structural elements are shown. ADP in EcHda and CDP in SaHda are shown in magenta and blue sticks, respectively. Each N-terminus of EcHda or SaHda is marked at the bottom with red or blue dots, respectively. (C) Close-up view of the nucleotide-binding site of the superimposed structures shows the interactions between nucleotide and hydrophobic residues of the lid domain. (D) The superimposed ADP and CDP of EcHda and SaHda, and the α-phosphate and β-phosphate marked by yellow or cyan dots are shown. The sticks are colored as in (B).