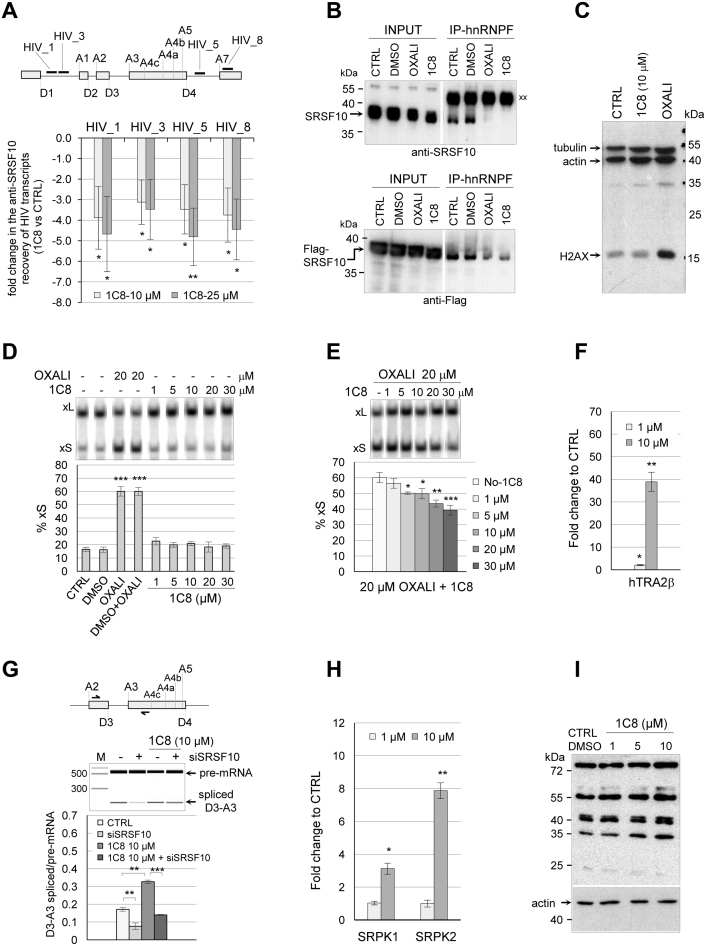
Figure 7.
1C8 affects the interaction of SRSF10 with HIV-1 transcripts and splicing factors. (A) The RNA recovered from anti-Flag immunoprecipation of extracts from HeLa-HIV cells treated or not with 1C8 was quantitated by RT-PCR using primers mapping to different portions of the HIV-1 pre-mRNA. The approximate position of amplicons on the HIV-1 genome is shown on top (HIV_1, HIV_3, HIV_5 and HIV_8). For each amplicon, the fold change between values in the 10 or 20 μM samples of 1C8 versus the non-treated control is plotted. (B) Immunoprecipitation of SRSF10 (top panel) and 3XFlag-SRSF10 (bottom panel) was performed with anti-hnRNP F antibodies using extracts prepared from non-treated 293 cells (CTRL) or treated with DMSO, 20 μM oxaliplatin, or 20 μM 1C8. The input content of SRSF10 and 3XFlag-SRSF10 in various samples is shown and represents 1/50th of the samples used for the immunoprecipitation. Immunoprecipitates were fractionated on gel and proteins were transferred to nitrocellulose that were decorated with anti-SRSF10 and anti-Flag antibodies. ‘xx’ indicates the large immunoglobulin subunit used for the immunoprecipitation that reacts with the secondary antibody. (C) Total proteins from control 293 cells and cells treated for 24 h with 20 μM 1C8 or 20 μM of oxaliplatin were fractionated by SDS-PAGE gel and transferred to nitrocellulose that was decorated with antibodies against phosphorylated H2AX, β-actin and tubulin. (D) The impact of 1C8 on Bcl-x splicing was tested by treatment of 293 cells for 24 h with the indicated concentrations of 1C8 and oxaliplatin. Total RNA was extracted and the Bcl-x splicing profile was revealed by RT-PCR. (E) The indicated concentrations of 1C8 were added to 293 cells one hour before adding 20 μM of oxaliplatin. After 24 h, total RNA was extracted and the percentage of Bcl-xS mRNA splice variant was determined. (F) The recovery of hTRA2β peptides from the anti-Flag immunoprecipitation of ribonuclease-treated samples was quantitated by LC–MS/MS analysis after normalization for equivalent of SRSF10 in all recovered samples. The presence of the hTRA2β peptides in the 1C8-treated samples was compared with control mock-treated samples. The mean fold change of recovered hTRA2β peptides is shown. (G) Impact of 1C8 and siSRSF10 on HIV-1 D3/A3 splicing in HeLa-HIV cells. Primer pairs indicated on top were used to perform endpoint RT-PCR, allowing to amplify both unspliced and spliced products. Analysis and visualization of amplified products were done using the LabChip HT DNA assay on a Caliper LC-90 automated microfluidic station. Typical electropherograms are shown, as well as histograms from an assay done in triplicate.(H) Similar to panel F, the mean fold change of SRPK1 and SRPK2 peptides recovered from 1C8-treated and mock-treated samples is shown in histograms. (I) Immunoblot with mAb104, which recognizes a phosphoepitope shared by the major SR proteins (51). The blot used gel-fractionated protein samples from HeLa cells treated for 18 h with the indicated concentrations of 1C8. In panels A and D–H, asterisks represent P values (two-tailed Student's t test) comparing the means between samples and their respective controls; *P < 0.05, **P < 0.01 and ***P < 0.001.