Fig. 1.
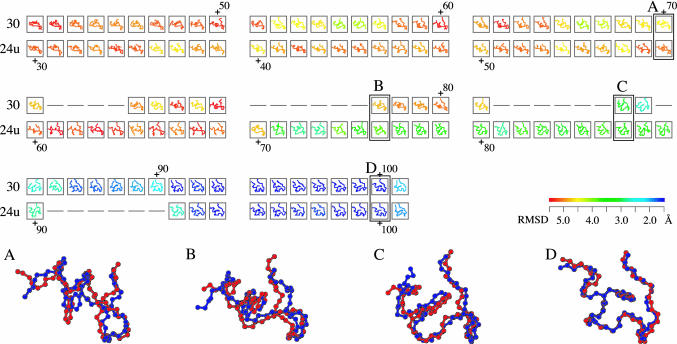
A part of the alignment of folding trajectory 30 and the time-reversed-unfolding trajectory 24u. All structures are colored according to rmsd of the NMR structure (21). The color gradient from red to blue indicates a decrease in rmsd from >5.5Åto <1.5 Å (see color bar). The similarity score S of the alignment is 27.3. The average rmsd between aligned 84 pairs of the snapshots is 2.2 Å. The magnified views of the aligned snapshots (A–D) show how similar these two folding pathways are; the blue and red chains are of trajectories 30 and 24u, respectively, whose rmsd values of A–D are 1.9, 2.2, 2.0, and 0.7 Å, respectively. Model 32 of PDB ID code 1l2y was used as the experimental structure, because it is closest to the average structure of the 38 models in 1l2y (21).