Figure 3. Prp modeled and docked with a substrate peptide.
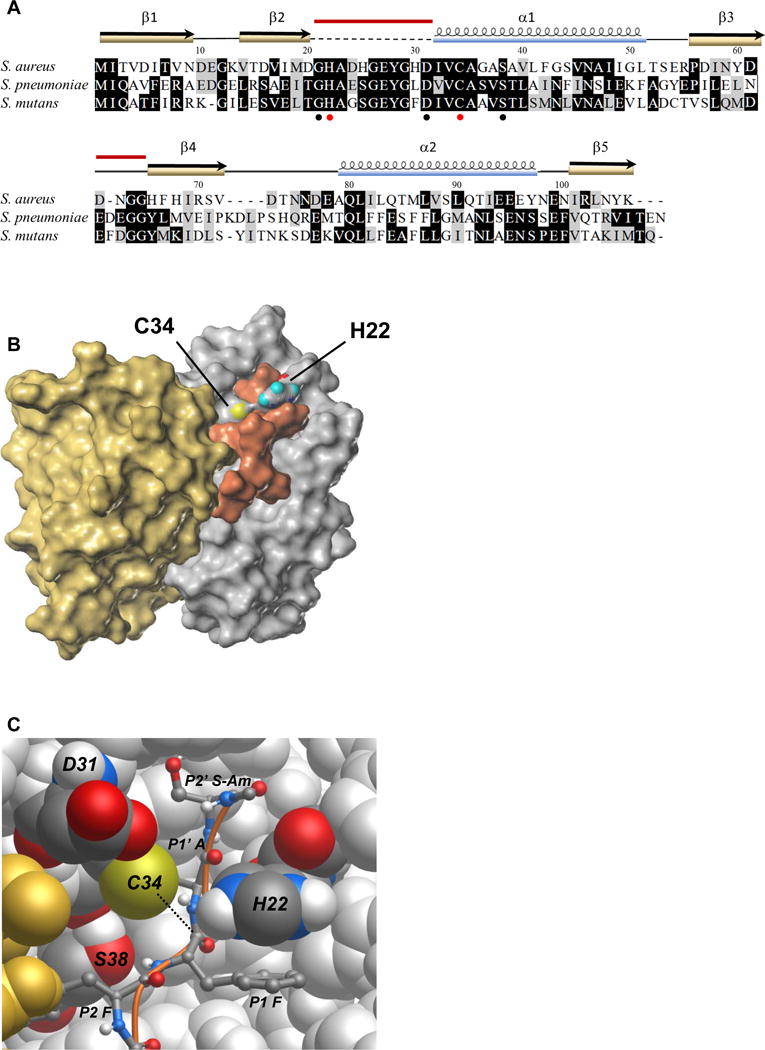
(A) Alignment of S. aureus Prp with the homologs from S. mutans and S. pneumoniae for which crystal structures have also been reported. Identical residues are highlighted in black, similar residues in gray. Conserved residues Gly 21, Asp 31, and Ser 38 surrounding the catalytic center are noted with black circles and the catalytic pair His 22 and Cys 34 are marked with red circles. The secondary structure elements from the crystal structure of S. aureus Prp are shown above the alignment. The dashed line indicates the region that was absent in the crystal structure, and the red bars indicate the two regions that were modeled in this study. PDB accession numbers: S. aureus (2P92/4PEO), S. mutans (2G0I), S. pneumoniae (2IDL). Amino acid sequences were aligned using MUSCLE and formatted in BOXSHADE through the Mobyle portal (Neron et al., 2009).
(B) Surface map of the Prp dimer and docked substrate Ac-NLQFFAS-Am. The catalytic pair residues are labeled and colored according to atomic convention (sulfur in yellow, oxygen in red, nitrogen in blue and hydrogen here in teal). Prp chain A is depicted in silver, chain B is in gold and the peptide substrate is shown in copper. In this model, the flexible loop that did not crystallize in PDB ID 2P92/4PEO is arranged in an arc above the active site. The substrate appears to associate with both chains of the enzyme.
(C) Atomic model of the Prp catalytic site. Prp chain B is in gold, chain A is silver with conserved labeled residues shown in atomic convention colors as above, with carbon shown in graphite and hydrogen in white. The peptide substrate Ac-NLQFFAS-Am is pictured as a ball and stick model with a copper backbone. In this depiction only the substrate residues FFAS-Am are visible (P2–P2′). The nucleophile (thiolate on Cys 34) is positioned to attack the carbonyl carbon of the scissile bond between P1 Phe and P1′ Ala on the substrate peptide (dotted line). The imidazolium group of His 22 is shown here π-π stacking with the phenyl group of the P1 Phe and, owing to its flexibility provided by Gly 21 (not visible), is positioned correctly to have abstracted a proton from Cys 34. The conserved hydroxyl group of Ser 38 is thought to provide a hydrogen bonding partner for the amide group between the P2 and P1 Phe residues. The role of Asp 31 is not clear in this model; its side chain protrudes near the catalytic site but appears to contact only solvent.
