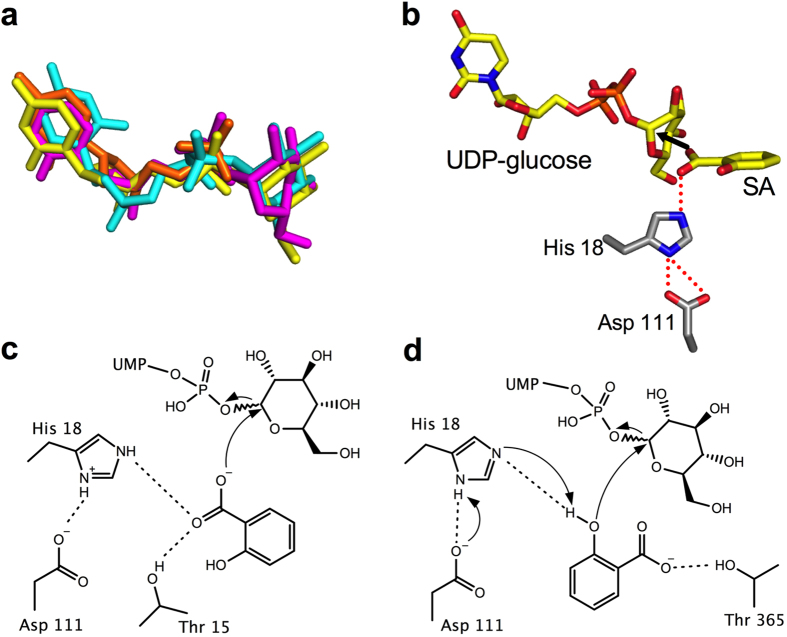
Figure 5. Proposed catalytic mechanism for SA glucose conjugate formation.
(a) Modeled UDP-glucose molecules. Crystallographically observed UDP shown in orange, UDP-2-fluoro-glucose from PDB 2c1z shown in yellow, UDP-glucose modeled by Molecular Operating Environment is in cyan, and UDP-glucose modeled from incomplete electron density is shown in magenta. (b) View of proposed catalytic site for UGT74F2 showing modeled UDP-glucose, SA and the dyad His 18 - Asp 111. (c) View of catalytic site to form SGE, based on (b) and Fig. 4a: SA carboxyl oriented towards UDP-glucose by interactions with His 18 and Thr 15, performs an SN2 attack by the carboxylate oxygen to the anomeric carbon of UDP-glucose. (d) View of catalytic site for SAG formation, based on Fig. 4e: SA hydroxyl deprotonated by His 18 - Asp 111 catalytic dyad; SN2 attack by the adjacent aromatic hydroxyl to the anomeric carbon results in the formation of SAG, and the proton is released from His 18 to regenerate the active site.