Abstract
The fight against cancer is hindered by its highly heterogeneous nature. Genome-wide sequencing studies have shown that individual malignancies contain many mutations that range from those commonly found in tumor genomes to rare somatic variants present only in a small fraction of lesions. Such rare somatic variants dominate the landscape of genomic mutations in cancer, yet efforts to correlate somatic mutations found in one or few individuals with functional roles have been largely unsuccessful. Traditional methods for identifying somatic variants that drive cancer are ‘gene-centric’ in that they consider only somatic variants within a particular gene and make no comparison to other similar genes in the same family that may play a similar role in cancer. In this work, we present oncodomain hotspots, a new ‘domain-centric’ method for identifying clusters of somatic mutations across entire gene families using protein domain models. Our analysis confirms that our approach creates a framework for leveraging structural and functional information encapsulated by protein domains into the analysis of somatic variants in cancer, enabling the assessment of even rare somatic variants by comparison to similar genes. Our results reveal a vast landscape of somatic variants that act at the level of domain families altering pathways known to be involved with cancer such as protein phosphorylation, signaling, gene regulation, and cell metabolism. Due to oncodomain hotspots’ unique ability to assess rare variants, we expect our method to become an important tool for the analysis of sequenced tumor genomes, complementing existing methods.
Author summary
The analysis of somatic variants in sequenced tumor samples is important for understanding the molecular disruptions that underlie the vast differences in individual cancer phenotypes or response to treatment. In order to understand which somatic mutations are functionally important for the initiation or progression of cancer, traditional analyses are ‘gene-centric’ in that they focus on single genes with high mutation frequency in tumor samples. However, many genes with experimental evidence of cancer involvement are found to be mutated in only a few tumor samples, hampering the data-driven identification of important genes. In our analysis, we leverage decades of important findings from structural genomics into the study of somatic variants by utilizing conserved protein domain families. Our method identifies ‘oncodomain hotspots’, sites within protein domain families with high mutation frequency in tumor samples. This enables our method to assess the importance of even rare variants by comparing to other genes with the same protein domain. By incorporating the structural and functional context encapsulated in protein domain families, we can identify even rare somatic variants in 5,437 genes, 3,041 of which are novel gene associations to cancer but are similar in structure and/or function to known cancer genes.
Introduction
In recent years, studies analyzing sequenced tumor genomes have seen a drastic increase in their sample sizes, growing from only a handful samples to cohorts of several thousand patients. This rise in availability of sequenced tumor samples has enabled the comparative analysis of tumors originating from different tissues, revealing a diverse tissue-specific genomic landscape of mutational patterns [1–6]. Revelations of this complexity observed in sequenced tumor samples has led to new insights into cancer genomics. However, identifying which somatic variants are the “drivers” behind initiation or progression of cancer is confounded due to the high prevalence of “passenger” mutations that occur with low frequency but are thought to have no functional effect [7,8]. Thus, despite the increase in tumor-derived data, we are unable to understand whether the vast majority of somatic variants in tumor samples have any functional role.
Towards understanding which somatic variants influence the initiation or progression of cancer, much work has been devoted to the cataloging of sequencing data in repositories like the Catalog of Somatic Variants in Cancer (COSMIC) [9] and to manually curated lists of genes with evidence of cancer involvement in GeneCards [10], the Cancer Gene Census (CGC) [11], the NCI Cancer Gene Index [12], the “proto-oncogene” and “tumor suppressor” classifications in the UniProt [13] database, the Network of Cancer Genes [14], and the TSGene database [15]. Massive ongoing sequencing projects like The Cancer Genome Atlas (TCGA) have discovered thousands of genes that are mutated in only a small fraction of tumors yet may still be important for cancer initiation or progression [7,16–18]. This has led to a rise in the availability of tools for analyzing and visualizing data [19–23] and also for identifying genes in tumor samples that are likely to harbor somatic variants that drive cancer initiation or progression [1,2,24,25]. Traditionally, methods for identifying important genes in tumor samples identify genes that are significantly enriched with somatic variants by clustering somatic variants by genes for statistical analysis. Clustering variants by gene regions is the natural choice since genes are units of inheritance and much is known about the function of particular genes. Not surprisingly, gene-centric studies of TCGA data have been able to recapitulate much of the knowledge about cancer genetics derived from decades of studies [1,2,6,24,25]. For instance, methods like the Cancer Mutation Prevalence Score (CaMP Score) in Sjöblom et al. [1], Wood et al. [2], and MutSigCV in Lawrence et al. [24] employ frequency-based analyses to identify regions of the genome (i.e., genes) that contain more mutations than expected by chance given a background of randomly occurring passenger mutations. However, the gene-centric analysis of individual cancer data relies on the relative frequency of all variants in a gene in sequenced tumor samples and is likely to miss variants that influence cancer progression that occur with relatively low frequency in the population. Even in the early years of such gene-centric data-driven analyses of sequenced tumor genomes like the CaMP Score, it was discovered that the genomic landscapes of somatic mutations in cancer were dominated by ‘gene hills’, or gene regions that are mutated at a low frequency. Indeed, it has been shown that even well-studied genes in cancer are mutated in only a small portion of tumor samples [18,26]. Thus, to identify infrequently mutated genes that play a role in cancer progression, other methods have been developed for clustering low frequency gene-mutations together with other genes with a common functional role. For example, clustering variants from genes on the same pathway [24,27–30], ontological term [28,31], or protein interacting partners [32,33]. Additionally, akin to tools for predicting deleterious variants in other diseases, machine learning methods [34–36] have been developed to determine which variants are likely to influence cancer progression. For instance, the Cancer-specific High-throughput Annotation of Somatic Mutations (CHASM) [34], is a machine learning predictor trained to classify between variants known to drive cancer progression and putatively neutral variants using properties of genomic and protein sequence, predicted protein structure, and multiple sequence alignments.
In recent work, Nehrt et al. [37] and Yang et al. [38] have shown the value of analyzing cancer somatic variants by clustering variants within a gene sub-region, i.e., the protein domain. Protein domains are the functional, structural, and evolutionary units of proteins [39,40], mediate approximately 75% of protein-protein interactions [41], and mutations in different domain regions of the same gene can have functionally and phenotypically distinct effects [42]. So, protein domain level studies have shown great potential to analyze tumor variants, in particular because they overcome the inability to distinguish functionally relevant variants due to the modularity and polyfunctionality of genes. In their domain-centric studies, somatic variants from TCGA of two [37] and later twenty [38] tumor types were analyzed to identify specific domain regions within genes that are significantly mutated in somatic tumor samples. In Nehrt et al., it was discovered that domain regions within a single gene can display heterogeneous mutation patterns that are unique between Breast Invasive Carcinoma and Colorectal Adenocarcinoma. Extrapolated to the plethora of cancer types available in the TCGA project, Yang et al. further defined these unique domain mutational patterns, highlighting patterns specific to any of these cancer types. In these previous domain-centric analyses, statistical measures were performed to identify domain families that are frequently mutated often with mutations from multiple genes with a common protein domain. In this work, we develop a novel method to identify “oncodomains”, or protein domains in which somatic variants from one or more genes encoding the domain occur more frequently at specific sites (i.e., oncodomain hotspots) than expected by chance. These oncodomain hotspots correspond to specific positions within an entire family of genes, which enables our method to study even extremely rare somatic variants via inference to other genes with similar somatic variant patterns. We argue that since protein domains are the structural and functional units of proteins, protein domains are the ideal framework for comparison to other genes since they are manually curated to match the structure and known functional features of domain family members, providing an inherent functional explanation of how somatic variants can contribute to cancer. To clarify, the approaches by Nehrt et al. and Yang et al. identified domain families that were enriched with somatic mutations but they did not, however, analyze the position-specific mutational patterns between different genes that share a common protein domain as in this work. The oncodomain concept introduced here is motivated by results from our earlier studies on known disease mutations. In Peterson et al. [43–45], we performed a domain-centric study to cluster all known disease variants into common domain regions from all human proteins. Results from these studies hinted at protein domain positions of functional relevance for the analysis of variants from the OMIM [46] and Swiss-Prot [47] databases. Specifically, known disease variants tend to cluster at specific domain sites more than expected by chance and these ‘position-based domain hotspots’ tended to be located on functional features and conserved residues, properties that were also found for variants that have been experimentally determined to be phenotypically altering in yeast [45]. Here we tested the hypothesis of whether cancer somatic variants also present similar patterns of aggregation as known disease variants. To address this question, we developed a new statistical framework in which we control for population-level frequency information and the large proportion of cancer passenger mutations. Oncodomain hotspots are derived exclusively from somatic mutations from sequenced tumor samples and represent a novel approach for assessing which somatic mutations are likely to influence the initiation or progression of cancer.
Although domain-centric models have been previously developed in Nehrt et al., Yang et al., the oncodomain method differs in substantial ways. Firstly, these studies were region-based in that entire domain regions were assessed for cancer significance, not specific positions within the domain family. Although Yang et al. identifies mutational hotspots, these hotspots are specific to a particular gene and contain no information from other genes sharing a common protein domain. Furthermore, the hotspots in Yang et al. do not consider variants from all domain regions as they restrict their analysis to domains that are significant in their region-based model. Secondly, oncodomains are inherently family-based in that somatic variants are aggregated to the domain-level and significance of a specific family member is ascertained by referencing all members of the family. Although Nehrt et al. analyzed domain regions from all genes sharing a common domain, the regions were concatenated and treated as a single, large gene and thus no positional information was used. Thirdly, the study conducted by Yang et al. only considers somatic variants that are predicted to be “potentially damaging” via the IntOGen-mutation platform [48] and removes all other somatic variants from the analysis. The IntOGen-mutation platform is a meta-predictor that classifies variants as “potentially damaging” primarily on the observed frequency in tumor samples and the results of several variant predictors, SIFT [49], PolyPhen-2 [50], VEP [51], and MutationAssessor [52]. This contrasts with oncodomain hotspots, which consider all somatic variants no matter the observed frequency and does not utilize machine learning methods to remove variants predicted to have no functional impact. Notably, filtering the data using variant predictors is problematic since it will bias the remaining variants towards conserved sites, functional features, structurally important residues, and even domain regions since this information is used in the variant predictors to assess deleteriousness.
In this work, we compare the results of oncodomain hotspots to genes with evidence of cancer involvement from the Cancer Gene Census, the NCI Cancer Gene Index, the Network of Cancer Genes, TSGene, and UniProt and to mainstream methods for the classification of cancer variants from tumors. Specifically, we compared to a gene-centric method, MutSigCV, two domain-centric approaches developed by Nehrt et al. and Yang et al., and a multi-feature machine learning predictor trained to distinguish drivers from passengers, CHASM. We demonstrate that oncodomain hotspots not only overlap well with the cancer genomics literature and the results of both gene- and domain-centric methods, but also that our method is unique in the ability to detect variants that occur with low frequency in tumor samples but have evidence of cancer involvement or are predicted to be driver mutations by CHASM. Due to the ability of oncodomain hotspots to leverage relevant structural and functional context to identify even rare somatic variants with high potential to drive cancer development, we hope for oncodomain hotspots to become an important tool for large-scale analysis of sequenced somatic tumor samples, complementing existing tools.
Materials & methods
Mapping somatic variants to specific protein domain positions
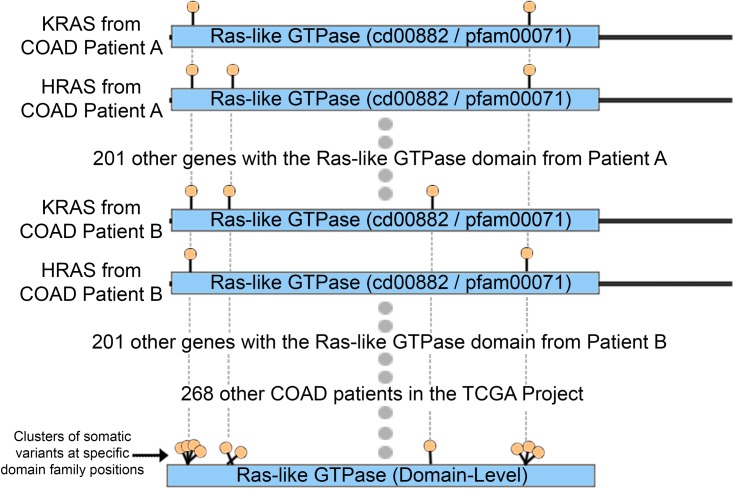
Somatic Variants from 5,848 patients from The Cancer Genome Atlas (TCGA) [53] were mapped to specific positions within protein domain models to identify clusters. TCGA MAF files were obtained on July 7th, 2014 for 20 cancer types: Adrenocortical Carcinoma (ACC), Bladder Urothelial Carcinoma (BLCA), Brain Lower Grade Glioma (LGG), Breast Invasive Carcinoma (BRCA), Colon Adenocarcinoma (COAD), Glioblastoma Multiforme (GBM), Head and Neck Squamous Cell Carcinoma (HNSC), Kidney Chromophobe (KICH), Kidney Renal Clear Cell Carcinoma (KIRC), Liver Hepatocellular Carcinoma (LHIC), Lung Adenocarcinoma (LUAD), Lung Squamous Cell Carcinoma (LUSC), Ovarian Serous Cystadenocarcinoma (OV), Pancreatic Adenocarcinoma (PAAD), Prostate Adenocarcinoma (PRAD), Rectum Adenocarcinoma (READ), Skin Cutaneous Melanoma (SKCM), Stomach Adenocarcinoma (STAD), Thyroid Carcinoma (THCA), and Uterine Corpus Endometrial Carcinoma (UCEC). Only validated exonic variants were used, resulting in 1,326,954 unique exonic variants across 20 cancer types. The number of patients and variants for each of the 20 cancer types studied is enumerated in Table 1. To map protein domain models to specific positions within human proteins, a human protein database containing 54,372 proteins was created with 33,963 proteins from RefSeq [54] and 20,409 proteins from Swiss-Prot [55] downloaded via NCBI’s E-utilities [56]. Since redundant protein entries exist between the RefSeq and Swiss-Prot databases, we selected only one representative protein for each unique Entrez gene ID, either the longest Swiss-Prot protein, or the longest RefSeq protein if no Swiss-Prot protein was listed for the gene ID. In addition, to avoid redundancy between isoforms produced by a single gene, we used only the longest protein product for analysis. Protein domain models from CDD [57] and Pfam [58] were obtained from the Conserved Domain Database (CDD version 2.25). HMMer’s semi global implementation [59] was used to map these domain models from human proteins. Finally, illustrated in Fig 1, proteins with somatic variants were aligned to specific positions within each domain model by using HMMer’s alignment with an E-Value threshold ≤ 0.001 where variants on gap regions of the domain model were assigned to the last position before the gap. To build the CDD protein domain set with minimal redundancy on the models, we selected only root domains (obtained from ftp://ftp.ncbi.nlm.nih.gov/pub/mmdb/cdd/cdtrack.txt). The final domain sets that map to human proteins contain 4,377 and 4,118 protein families from CDD and Pfam respectively.
Table 1. Number of patients, somatic variants, oncodomains, and oncodomain hotspots for each cancer type.
| Cancer Type | Number of Patients | Number of Exonic Somatic Variants | Number of Pfam Oncodomains (fdr(t) = 0.05) | Number of Pfam Oncodomains (fdr(t) = 0.01) | Number of Pfam Oncodomain Hotspots (fdr(t) = 0.05) |
Number of Pfam Oncodomain Hotspots (fdr(t) = 0.01) |
|---|---|---|---|---|---|---|
| ACC | 91 | 41,451 | 44 | 42 | 67 | 62 |
| BLCA | 130 | 39,312 | 31 | 17 | 73 | 31 |
| BRCA | 977 | 90,490 | 68 | 41 | 255 | 123 |
| COAD | 270 | 125,522 | 148 | 108 | 646 | 435 |
| GBM | 291 | 22,166 | 32 | 21 | 69 | 47 |
| HNSC | 306 | 74,008 | 4 | 3 | 7 | 6 |
| KICH | 66 | 3,835 | 1 | 1 | 1 | 1 |
| KIRC | 422 | 55,092 | 52 | 36 | 139 | 70 |
| LGG | 289 | 14,817 | 20 | 17 | 45 | 33 |
| LIHC | 202 | 92,840 | 74 | 49 | 364 | 182 |
| LUAD | 542 | 255,972 | 142 | 77 | 1,550 | 1212 |
| LUSC | 138 | 49,997 | 30 | 21 | 210 | 116 |
| OV | 375 | 21,207 | 16 | 10 | 49 | 43 |
| PAAD | 91 | 46,505 | 41 | 29 | 73 | 42 |
| PRAD | 259 | 9,437 | 7 | 6 | 18 | 11 |
| READ | 116 | 34,259 | 52 | 31 | 106 | 67 |
| SKCM | 344 | 290,341 | 345 | 200 | 1,742 | 1,258 |
| STAD | 289 | 148,520 | 119 | 78 | 828 | 574 |
| THCA | 402 | 7,458 | 8 | 7 | 12 | 7 |
| UCEC | 248 | 240,546 | 258 | 161 | 1,547 | 1,186 |
Fig 1. Depiction of the process of mapping variants to domain positions to find oncodomain hotspots.
Identifying cancer-specific oncodomain hotspots within protein domain families
In previous work by Peterson et al. [43,44] and Yue et al. [60] it was shown that variants with known cancer relevance from the OMIM and UniProt databases tend to cluster at positions within protein domains. However, the inclusion of patient frequency information is critical for the analysis of TCGA somatic variants from sequenced tumor samples and for the identification of driver mutations, but requires a new statistical framework that includes patient frequency into the analysis. Thus, in this work, we developed a mutational score to classify protein domain positions derived from individual patient data using a local false discovery rate (FDR) with a Zero-Inflated Poisson (ZIP) null distribution. We applied this methodology separately for each cancer type and for each protein domain model and defined high scoring protein domain positions as those with a q-value < 0.05. The details and derivation of this statistical approach can be found in a separate work by Gauran et al. [61] but briefly, the formulation used is as follows.
At the protein domain-level which often encompasses several genes, each position within the domain contains j = 0,1,…jmax somatic mutations from patients with the same cancer type and we define nj as the number of domain positions with j somatic variants. We developed a local false discovery rate method using a zero-inflated Poisson distribution as the null distribution for non-significantly mutated positions. Each protein domain was considered separately to remove the influence of region-based cofactors (replication timing, expression, etc.) since each domain position is aligned to the same set of proteins. Our goal is to find the cutoff of j which separates non-significantly (f0(j)) and significantly (f1(j)) mutated positions. The observed count of mutations are from a mixture distribution, where
Where f0 is assumed to follow a Zero Inflated Poisson (ZIP) distribution while f1 could be any other (discrete) distribution. ZIP models are considered useful for the analysis of count data with a large amount of zeros because it allows for two sources of overdispersion by mixing a Poisson distribution with zero-inflation. For a given position, we assume that the number of mutations j is generated by one of the two distributions f0(j) or f1(j) so the probability density function of the mixture distribution is
Then, we define the local FDR at t as
Which indicates that f dr(t) is the posterior probability that a position with j = t is non-significant. The interpretation of the local FDR value is analogous to the frequentist’s p-value wherein local FDR values less than a specified level of significance provide stronger evidence against the null hypothesis. In this work, unless noted otherwise, we use a cutoff of f dr(t) = 0.05, which would indicate that only 5% our oncodomain hotspots are false discoveries.
When comparing regions of the genome (i.e., genes in the CaMP score and MutSigCV), methods must account for “covariates” that are thought to influence the background rate of passenger mutations for that particular genomic region, such as replication timing, gene expression, chromatin state (open/closed), and mutation context (e.g., C to G in CpG sites, G to C in GpA sites, etc.). When analyzing an aligned position within the same family of genes, the altered mutation rate of the aligned gene regions does not differ between aligned positions and thus does not need to be modeled. This is correct for all covariates with the exception of mutational context, which may differ between aligned positions. However, we determined that using synonymous variants to estimate the background probability of passenger mutations was inappropriate. Firstly, it is well known that many synonymous variants are drivers that re-occur in cancer and are not distributed randomly [62–64]. Secondly, the frequency of occurrence of synonymous variants is often different than that of the nonsynonymous variants, making them inappropriate to use to estimate the null model. Thus, using a randomly distributed background of equal size to the observed nonsynonymous variants was chosen.
Overlap with functional features and conserved positions
To assess the significance of overlap between oncodomain hotspot positions and positions that have known function, functional feature annotations for each protein position were obtained from UniProt on July 18th 2015. To determine the conservation of each domain position j, we employed the AL2CO [65] algorithm for assessing entropy via the following formula:
Here, p(ai,j) is the amino acid frequency for amino acid ai at position j and Hj is the AL2CO score at position j. Positions were considered to be conserved if they were greater than or equal to the average AL2CO score plus one standard deviation. Pearson’s correlation coefficient and Fisher’s exact test with Bonferonni correction were used to assess significance of hotspot position overlap with functional features or conserved residues.
Comparison to other methods & cancer-related databases
To compare to other methods, significantly mutated genes were obtained using MutSigCV v1.4, significantly mutated domains were obtained from the results of Nehrt et al. and Yang et al., and the results of CHASM were obtained from the Firehose project [19]. To compare to cancer-related databases, the Gene Ontology database [66] along with the pfam2go annotations were obtained on August 21st 2015, the NCI Cancer Gene Index was obtained on March 7th, 2016, the Network of Cancer Genes was obtained on March 4th, 2016, and the TSGene database was obtained on March 4th, 2016, the Cancer Gene Census [11] on November 6th, 2015, and the UniProt [13] “proto-oncogene” and “tumor suppressor gene” classifications were obtained on November 7th, 2015. Gene Ontology category enrichment was performed using Fisher’s exact test with Bonferroni correction.
Results
Oncodomains and cancer-specific oncodomain hotspots
In this work, we define oncodomains as families of protein domains in which somatic variants from one or more genes containing the same domain form a hotspot. Oncodomain hotspots are defined as protein domain positions where somatic variants for a specific cancer type occur more frequently than expected by chance (see Materials & Methods). A comparison of the number of oncodomains and oncodomain hotspots identified for different fdr(t) cutoffs along with the number of patients and exonic somatic variants for each cancer type is shown in Table 1. For simplicity, we will refer to the results obtained using the fdr(t) cutoff of 0.05 for the remainder of this analysis. In this study, we identify 185 protein domain families from CDD and 673 from Pfam across 20 cancer types as oncodomains. Within these families, 2,126 oncodomain hotspots were identified on CDD domains and 3,563 hotspots were identified on Pfam domains. Overall, the quantity and location of the hotspots were found to be highly heterogeneous between cancer types. We find the number of oncodomains and oncodomain hotspots to be highly variable between cancer types ranging from only 1 or 7 hotspots in KICH and HNSC respectively, to a maximum of 1,742 hotspots identified in SKCM. In our dataset, TCGA cancer types had an average of 74 (standard deviation of 89.8) oncodomains and an average of 309 (standard deviation of 571) oncodomain hotspots. The frequency of hotspots across the 20 cancer types was highly heterogeneous with nearly 400 domain models being signatures for only one cancer type while 21 were common to ten or more cancer types (S1 Fig & S1 File). A full list of all oncodomains and the cancer-specific oncodomain hotspots for each cancer type can be found in S2 File.
We find a strong correlation between the total number of exonic somatic variants and the number of oncodomains / oncodomain hotspots (Pearson’s Correlation 0.92 and 0.98 respectively). Compared to the number of exonic variants, the number of patients in each cancer type was not as strongly correlated to the number of oncodomains (Pearson’s Correlation: 0.14) or oncodomain hotspots (Pearson’s Correlation: 0.21), which is to be expected since the number of somatic variants per tumor is known to be highly variable between cancer types [25]. However, the importance of including more sequenced patients for research is highlighted in S1 Table. To address this, a bootstrapping analysis was performed 100 times for the three largest TCGA sets (LUAD, SKCM, and UCEC) to calculate oncodomains and oncodomain hotspots using only 75% and 50% of the available patients and, separately, the available exonic somatic variants. Results for bootstrapping patients or variants both suggest that more oncodomains and oncodomain hotspots will be identified when more data become available, as expected.
We also tested the effect of combining patients from all cancer types to observe whether oncodomains and oncodomain hotspots differ from the cancer-specific hotspots analysis. In this separate analysis, we observe an increase of 82 oncodomains and 1,469 oncodomain hotspots (Pfam only) when combining all data types together that were not identified when analyzing the sets individually (S3 File). Results from the combined dataset also show that 247 oncodomains and 1,251 oncodomain hotspots that were previously identified when analyzing individual datasets are no longer significant in the combined dataset. This, however, is to be expected due to the disproportionate number of patients in each cancer type, removing much of the cancer-specific signals.
Cancer-specific heterogeneity in oncodomain family somatic mutation rates
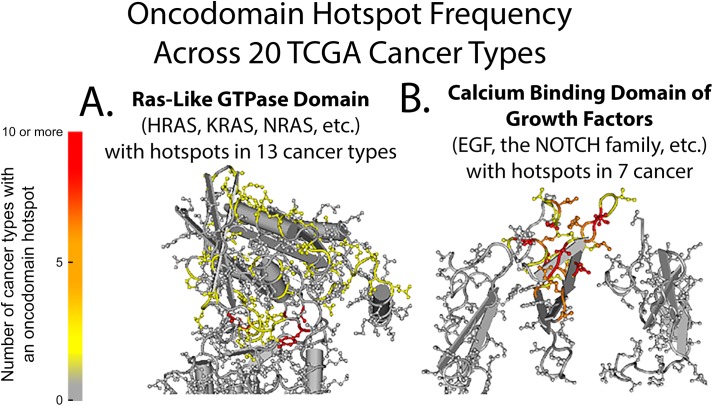
Like genes, protein domains have been shown by Nehrt et al. and Yang et al. to display heterogeneity in the prevalence of somatic variants from patients with different cancer types. However, no study yet has explored the mutation patterns of domain families that appear several times throughout the human genome. In our analysis, we observed this heterogeneity in the prevalence of somatic variants between different cancer types and also between the frequencies in which members of a particular domain family are involved. For example, in S2 File, the hotspots formed on a particular oncodomain are found to be highly heterogeneous in the quantity and location for a given cancer type. Depicted in Fig 2 for the Ras-like GTPase family (Fig 2A) and the calcium binding domain of the Epidermal Growth Factor (Fig 2B), the intensity of color at each residue represents the number of cancer types in which that residue was found to be an oncodomain hotspot across the 20 cancer types. In these structural representations, the frequency or specific location in which somatic variants occur is highly heterogeneous between cancer types, a property that would normally be ignored by traditional region-based analyses that group all positions within a gene or domain region into a single bin when testing for significance.
Fig 2. Hotspot frequency of the Ras-like GTPase oncodomain and the calcium binding Epidermal Growth Factor domain.
Structural representations of the Ras-like GTPase (cd00882) oncodomain family (A) and the calcium binding domain of the epidermal growth factor-like (cd00054) oncodomain family (B).
Enrichment of functional features, conserved residues, & ontological terms
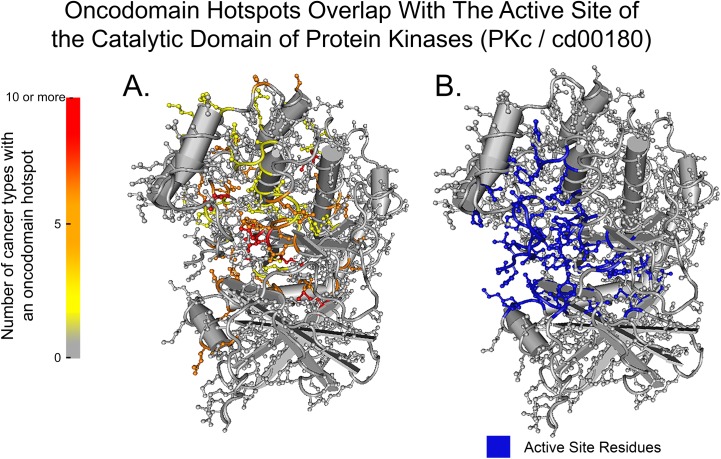
The overlap between oncodomain hotspots and functional features for each protein residue in the UniProt database were ranked by their Fisher’s exact test p-value with Bonferroni correction and are listed in Table 2. Overall, we found that oncodomain hotspots significantly occur on functional feature sites (p-value: 3.63E-87), a finding that is not true for somatic variants overall, which do not occur significantly at functional feature sites (p-value > 0.05). Interestingly, the specific residue of the functional feature that is mutated is heterogeneous between cancer types, as seen in the comparison between the frequency of mutated sites in Fig 3A and the residues involved with the active site in Fig 3B. Additionally, we found a significant overlap between oncodomain hotspots and conserved residues (p-value: 1.45E-09). However, conservation and functional feature annotation do not correlate with oncodomain hotspots (Pearson’s correlation coefficients of 0.009 and 0.048 respectively), indicating that this information alone is insufficient for determining which functional or conserved residues will be important for cancer initiation or progression. For genes with a somatic variant in an oncodomain hotspot, enrichment was performed for categories of genes in the Molecular Function and Biological Process divisions of the Gene Ontology database (S2 Table). For Pfam oncodomains, Gene Ontology term enrichment was performed using the pfam2go annotations (S3 Table).
Table 2. Enrichment of residues with functional feature annotation.
| Hotspot Enrichment for Functional Feature Annotation on Protein Positions | |
|---|---|
| Feature Name | P-Value |
| Nucleotide binding site | 2.8E-155 |
| DNA binding site | 3.3E-141 |
| Calcium binding site | 2.5E-11 |
| Active site | 1.7E-7 |
| Metal binding site | 3.5E-3 |
Fig 3. Overlap of oncodomain hotspots with the active site of the catalytic domain of protein kinases.
Structural representation of the frequency of oncodomain hotspots across 20 cancer types (A) compared to the active site residues (B) for the PKc / cd00180 oncodomain.
Comparison to other methods & databases
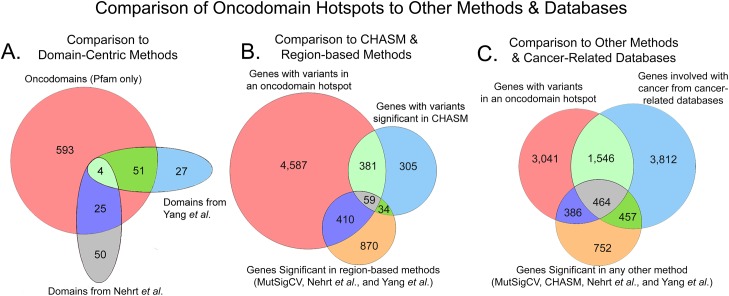
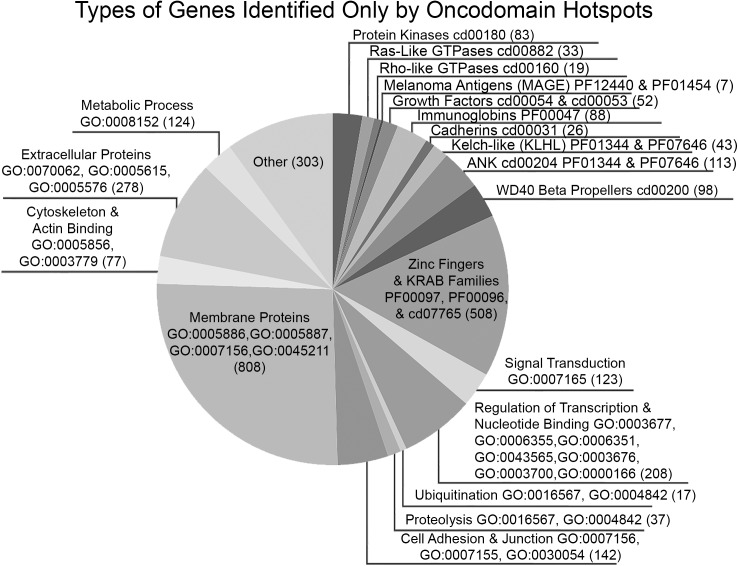
Overall, we found that oncodomain hotspots identify more protein domains, genes, and somatic variants than other methods, many of which are rare variants. Due to the lack of a good benchmarking set, we compared the results of our method to the results of other methods for analyzing somatic tumor genomes and to databases of genes with evidence of cancer involvement. In comparison to other domain-centric methods (Nehrt et al. and Yang et al., Fig 4A), oncodomain hotspots recapitulate 80 / 157 (51%) of Pfam domain models while identifying 593 novel Pfam models. At the gene-level in Fig 4B, genes with variants in an oncodomain hotspot identify 440 / 779 (56%) of genes with variants significant in CHASM, 469 / 1,373 (34%) of genes identified by region-based methods (MutSigCV, Nehrt et al., and Yang et al.), and 4,587 genes were unique to oncodomain hotspots. Of these 4,587 genes unique to oncodomain hotspots, we found 1,546 / 4,587 (34%) genes to have evidence of cancer involvement from the Cancer Gene Census, the NCI Cancer Gene Index, the Network of Cancer Genes, the Uniprot “proto-oncogene” and “tumor suppressor gene” classifications, and the TSGene databases (Fig 4C) which were not detected by MutSigCV or CHASM. As depicted in Fig 5, the majority of the remaining genes detected only by oncodomain hotspots (2,738 / 3,041; 90%) are either members of domain families for which cancer relevance is known (e.g., kinases, growth factors, and immunoglobins) or are annotated with GO terms that have known cancer relevance (e.g., signal transduction, metabolic process, and cell adhesion).
Fig 4. Comparison of oncodomain hotspots to other methods and databases.
Fig 5. Types of genes identified only by oncodomain hotspots.
Oncodomain hotspots enable the functional analysis of rare somatic variants
Rare variants are thought to play an important role in cancer and, thus, frequency-based methods are inherently ill-suited to assess their relevance in cancer due to their low prevalence in tumor samples. However, by comparing to other genes within the same domain family, oncodomain hotspots have the ability to infer functional relevance of variants that occur infrequently in tumor samples. Indeed, variants implicated only by oncodomain hotspots occurred in an average of 1.1 (variance of 0.34) tumor samples compared to variants implicated by MutSigCV that occurred in an average of 2.1 (variance of 64.4) tumor samples (t-test p-value: 3.5E-259). On the other hand, as expected, oncodomain hotspots implicate many of the frequently occurring variants that would be identified by other methods since the variants in oncodomain hotspots that were also identified by MutSigCV occur in an average of 2.2 (variance of 59.3) tumor samples.
Discussion
Distinguishing between drivers and passengers in sequenced tumor samples is a challenging task in cancer biology. However, traditional methods that rely solely on frequency of somatic variants for identifying driver variants are limited due to the lack of sequenced patients, even with the thousands of patients that have been sequenced in TCGA. As noted in Sjöblom et al. and Wood et al., the genomic landscapes of somatic mutations are dominated by “gene hills”, or infrequently mutated genes that do not reach statistical significance but may still be relevant in cancer. Thus, new methods are needed in order to functionally characterize these rare variants and their importance in cancer. As shown in previous studies, Nehrt et al. and Yang et al., domain-centric analyses have the potential to identify somatic mutational patterns unique to specific cancer types that would normally be overlooked by gene-centric analyses that consider only whole proteins and not the modular regions within. Such approaches can help improve our understanding of the molecular perturbations leading to cancer initiation and progression and enable the identification of new targets for cancer-specific drug research. However, these approaches consider only variation between domain regions within a single gene and, as such, ignore similar, often rare variants in other members of the same protein family that may play a similar role in cancer or may also affect drug treatments. In this study, by leveraging the knowledge of conserved regions of proteins that can occur several times throughout the genome (i.e., protein domains), we are able to infer functional and structural relevance of rare somatic variants by comparing them to similar variants in other genes sharing a common protein domain. This novel concept also allows us to observe heterogeneity in mutation prevalence between members of a protein family—patterns which can be unique for particular cancer types.
In this work, we identify “oncodomain hotspots”, or positions within protein domain regions that harbor more somatic variants than expected by chance by aligning similar domain regions from multiple genes across all patients for a given cancer type (Fig 1). Overall, we found the location and intensity of oncodomain hotspots to be highly heterogeneous between cancer types. For example, as enumerated in S2 File, we found that position five on the Ras-like GTPase (Fig 2A) was the most frequently occurring hotspot on cd00882, appearing in 10 cancer types (BLCA, BRCA, COAD, LUAD, OV, PAAD, READ, SKCM, STAD, and UCEC) and represents a portion of the GTP/M2+ binding site. However, this hotspot was not found in THCA, where oncodomains identified, instead, a hotspot on position 307. Similarly, in LIHC, oncodomain did not identify position five or 307 as hotspots but we reported a hotspot at seven other positions, two of which can only be found in LIHC. Thus, some hotspot patterns are common in several cancers while others are unique to a specific cancer type. In the Ras-like GTPase alone, we find one hotspot unique to COAD, two hotspots unique to LIHC, five hotspots unique to LUAD, six hotspots unique to SKCM, three hotspots unique to STAD, and 20 hotspots unique to UCEC. Interestingly, while we observe a stark heterogeneity between the location and intensity of oncodomain hotspots between cancer types, our results show a significant overlap for oncodomain hotspot location with conserved residues and functional feature sites. Thus, although oncodomain hotspots are heterogeneous, they tend to occur at different positions that are highly conserved residues or at different positions that perform similar functions as seen in Fig 3 where hotspots tend to occur spatially around the active site of the catalytic domain of protein kinases.
Overall, oncodomain hotspots identify many more domains (Fig 4A) than other domain-centric methods like Nehrt et al. and Yang et al. and more genes (Fig 4B) than gene-centric methods like MutSigCV or CHASM. Although not identified by other methods, 1,546 / 4,629 (34%) of genes identified only by oncodomain hotspots have evidence of cancer involvement from the Cancer Gene Census, the NCI Cancer Gene Index, the Network of Cancer Genes, the Uniprot “proto-oncogene” and “tumor suppressor gene” classifications, and the TSGene manually curated databases (Fig 4C). Interestingly, we find variants in oncodomain hotspots on 392 genes from either the TSGene database or UniProt’s tumor suppressor gene annotations, indicating that both oncogenes and tumor suppressors form hotspots at the domain-level, a phenomenon previously discovered for tumor suppressor genes at the gene-level [67–69]. Moreover, as illustrated in Fig 5, the majority (90%) of the remaining 3,041 genes in Fig 4C identified only by oncodomain hotspots are either members of domain families for which cancer relevance is known or are annotated with GO terms that are known to be important for cancer. Overall, oncodomain hotspots find many new genes that display similar somatic variant patterns to other genes within the same domain family that are well-studied in cancer genomics including 83 novel kinases (cd00180), 52 novel growth factors (cd00054 & cd00053), 33 novel Ras family members (cd00882), 26 novel cadherins (cd00031), 88 novel immunoglobins (pfam00047), and 43 novel Kelch-like (KLHL) genes. Additionally, oncodomain hotspots identify significant somatic variant clusters in the Melanoma Antigen (MAGE) family of genes which were never significant in other methods as well as the Rho-like GTPase family, which has known cancer involvement but is notorious for being somatically mutated only rarely [70,71]. Oncodomain hotspots also identify many genes involved with cell adhesion and cell junction organization, which are known to be important in cancer progression [72–74] and metastasis [75,76], and genes involved with metabolism, which are also important in cancer progression [77–79]. Furthermore, many genes involved with the extracellular matrix or extracellular vesicles formed oncodomain hotpots, which are thought to be important in the regulation of cancer progression and metastasis [80–85]. Oncodomain hotspots are also formed on other gene families involved with processes thought to influence cancer initiation or progression such as ubiquitination [86–88], proteolysis [89–91], metabolic proteins [92,93], and genes involved with actin binding and the cytoskeleton [94–96]. Interestingly, oncodomain hotspots also identify many membrane proteins, which are involved with signal transduction, which is known to be relevant in cancer [97,98] and experimental evidence confirms the important regulatory role played by membrane proteins in cancer [99–105]. Our results also indicate a strong pattern of variants occurring at specific domain family sites for genes involved with signal transduction, regulation of transcription, and nucleotide binding GO terms. Likewise, we find oncodomain hotspots in domain families that serve as the molecular machinery of transcription factors (zinc fingers, KRAB domains, and WD40 beta propellers) as well as ANK domains, which mediate protein-protein interactions [106]. Thus, oncodomain hotspots reveal a vast landscape of somatic variants that act at the level of domain families altering signaling pathways and gene regulation to influence cancer.
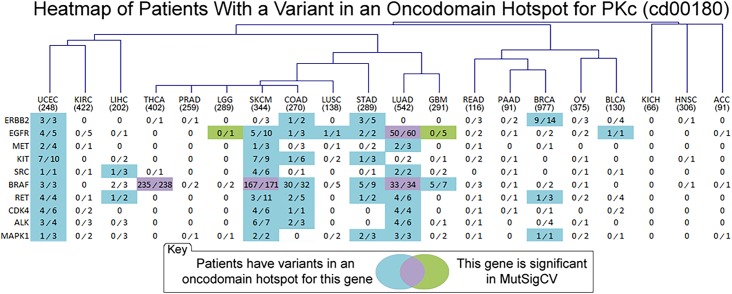
Identifying the role in cancer, if any, of so-called “gene-hills” in Sjöblom et al. and Wood et al. has been an important challenge since rare variants are thought to play an important role in cancer [6,107,108], which has led to an increase in network-based analyses for functional characterization [24,27–30]. A domain family-based analysis like oncodomain hotspots enables the identification of many novel, often rare variants that occur more frequently in specific positions within domain families than expected by chance. Indeed, when analyzing entire families of proteins and not specific members therein, mutational patterns emerge which suggest that rare variants play an important role since they often occur on genes with known cancer relevance. For example, protein kinases harbor somatic variants in 3,634/5,848 (62.1%) of the tumors analyzed in this study yet only 27 / 465 human genes mapping to the PKc (cd00180) domain model were considered significant by MutSigCV, 16 of which were significant in only the PAAD cancer type. In Fig 6 and S2 Fig, we summarize the results of comparing MutSigCV and CHASM respectively against oncodomain hotspots to evaluate the ability of these methods to identify rare and common variants relevant to cancer. The genes selected are members of the PKc (cd00180) oncodomain family, the catalytic domain of protein kinases that are the most frequently mentioned in PubMed articles annotated with the “cancer” MeSH term, effectively ranking them by how frequently they are mentioned in the cancer literature. This family contains 465 genes encompassing all serine-threonine, tyrosine, and dual specificity kinases in the human genome. Results in Fig 6 highlight the importance of rare variants in cancer since many genes with known cancer relevance are not reported by MutSigCV (shown in blue). Several instances exist where these MutSigCV and oncodomain hotspots agree (purple) and also where MutSigCV finds significance where the oncodomain method did not (green). Surprisingly, MutSigCV performed poorly for these genes since only two of these genes (EFGR and BRAF) were significant in MutSigCV for any cancer type. When compared to both MutSigCV and CHASM (S2 Fig), oncodomain hotspots still identify many more variants than MutSigCV and CHASM combined. However, CHASM is a machine learning method and does not incorporate the frequency of the variant but instead utilizes 70 features calculated from properties of genomic and protein sequence, predicted protein structure, and multiple sequence alignments. CHASM’s Random Forest algorithm is trained on a set of known driver mutations as a positive set and synthetically generated passenger mutations as a negative set. Thus, while MutSigCV would not be able to implicate these rare variants due to insufficient population frequency, CHASM uses properties learned from known driver mutations, which often agree with oncodomain hotspots that utilize population frequency alone. Furthermore, we find that oncodomain hotspots are capable of identifying more rare variants in these kinases than other methods while still identifying the obvious variants that occur with high frequency such as EGFR in LUAD and BRAF in THCA, SKCM, and LUAD. Moreover, oncodomain hotspots are able to identify genes that are known to be associated with particular cancer types where traditional methods may fail. For example the seven genes identified by oncodomain hotspots for COAD (ERBB2 [109,110], EGFR [111,112], KIT [113,114], BRAF [115–117], RET [118,119], CDK4 [120–122], ALK [123–125], and MAPK1 [126–128]) are reported to have been involved with COAD. Interestingly, all of these genes were found to be mutated in only six or fewer patients with the exception of BRAF, which was mutated in 32 patients but was still not identified by MutSigCV or CHASM in S2 Fig. In other examples, the SRC gene is a well-known oncogene involved in the PI-3K cascade but no other method is able to detect any significance while oncodomain hotspots identify 8 somatic variants in oncodomain hotspots for LIHC, LUAD, SKCM, and UCEC where some evidence of SRC’s role is known [129–131]. Even for genes that were significant in MutSigCV, oncodomain hotspots are more sensitive as they identify those same genes as significant in more cancer types for which they are known to play a role like BRAF in STAD [132–134], GBM [135–137], and UCEC [138,139] and EGFR in COAD [111,112], STAD [140,141], and SKCM [142–144]. Indicating the ability of oncodomain hotspots to implicate rare variants, 48 variants on these PKc genes that were found in three or fewer tumor samples fell into oncodomain hotspots and five of these variants were found in only a single tumor sample.
Fig 6. Heatmap of Patients with a Variant in an Oncodomain Hotspot for the PKc domain.
Visual representation and hierarchical clustering of oncodomain hotspots on genes that were significant in MutSigCV. For each gene in each cancer type, the number of patients in oncodomain hotspots is quantified and the cell is color-coded if the gene had any patients in oncodomain hotspots (blue), if it was significant in MutSigCV (green) or both (purple). Only the top ten genes based on the gene name’s co-occurrence with the “cancer” MeSH term are shown. Here, cancer types are grouped via hierarchical clustering to show similar mutational patterns. Enumerated in each cell are the proportion of patients with a somatic variant in an oncodomain hotspot (numerator) compared to the number of patients that had a somatic variant anywhere in the protein domain region (denominator).
To conclude, in sequenced tumor samples, even somatic variants that are known to drive tumor progression can occur with relatively low frequency. Our novel oncodomain method for identifying likely driver variants reveals the structural and functional mutational patterns on conserved protein domains that are unique to each cancer type. This allows us to infer functional importance of even rare somatic variants via inference to somatic variants in other genes sharing a common protein domain. Determining which variants are most important for tumorigenesis will help elucidate the mechanisms driving tumor progression and could ultimately provide a new set of drug targets for families of genes that display similar variation at the structural and functional level. We expect oncodomain hotspots to be an integral tool for assessing novel rare variants in tumor samples, complimenting other existing tools.
Supporting information
Frequency distribution of the number of times pfam oncodomain families form a hotspot in 20 different cancer types.
(TIF)
Visual representation and hierarchical clustering of oncodomain hotspots on genes that were significant in CHASM or MutSigCV. For each cell, the ratio of patients with somatic variants in a hotspot to patients with a somatic variant in the domain region is quantified. Each cell is color-coded if the gene had any somatic variants of that cancer type in an oncodomain hotspot (blue), if it was significant in CHASM/MutSigCV (green), or both (purple). Only the top ten genes based on the gene name’s co-occurrence with the “cancer” MeSH term are shown. Here, cancer types are grouped via hierarchical clustering to show similar mutational patterns. Enumerated in each cell are the proportion of patients with a somatic variant in an oncodomain hotspot (numerator) compared to the number of patients that had a somatic variant anywhere in the protein domain region (denominator).
(TIF)
Bootstrap analysis was performed to count the number of Pfam oncodomains and oncodomain hotspots with only 75% or 50% of the available patients or available exonic somatic variants. The bootstrapping process was repeated 100 times for each cancer type, bootstrap percentage, and local false discovery rate cutoffs.
(DOCX)
Enrichment of the Biological Process and Molecular Function Gene Ontology ontologies for genes with at least one somatic variant in an oncodomain hotspot for any cancer type.
(DOCX)
Top twenty enriched Gene Ontology terms with Pfam oncodomains from the pfam2go annotations using Fisher’s exact test with Bonferroni correction.
(DOCX)
(XLSX)
(ZIP)
(XLSX)
Acknowledgments
The authors would like to acknowledge Ann Cirincione for her valuable help in creating the figures. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found at http://cancergenome.nih.gov/.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by NSF (award #1446406, PI: MGK), NIH (award #1K22CA143148, PI: MGK and Award #R01LM009722 CoPI: MGK). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314: 268–274. 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- 2.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113. 10.1126/science.1145720 [DOI] [PubMed] [Google Scholar]
- 3.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, et al. (2009) Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature 462: 1005–1010. 10.1038/nature08645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, et al. (2011) The mutational landscape of head and neck squamous cell carcinoma. Science 333: 1157–1160. 10.1126/science.1208130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson IR, Takahashi K, Futreal PA, Chin L (2013) Emerging patterns of somatic mutations in cancer. Nat Rev Genet 14: 703–718. 10.1038/nrg3539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parmigiani G, Lin J, Boca S, Sjoblom T, Kinzler K, et al. (2007) Statistical methods for the analysis of cancer genome sequencing data.
- 7.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158. 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stratton MR, Campbell PJ, Futreal PA (2009) The cancer genome. Nature 458: 719–724. 10.1038/nature07943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, et al. (2011) COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res 39: D945–950. 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, et al. (2010) GeneCards Version 3: the human gene integrator. Database (Oxford) 2010: baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, et al. (2004) A census of human cancer genes. Nat Rev Cancer 4: 177–183. 10.1038/nrc1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alliance SS NCI Cancer Gene Index.
- 13.(2015) UniProt: a hub for protein information. Nucleic Acids Res 43: D204–212. 10.1093/nar/gku989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.An O, Dall'Olio GM, Mourikis TP, Ciccarelli FD (2016) NCG 5.0: updates of a manually curated repository of cancer genes and associated properties from cancer mutational screenings. Nucleic Acids Res 44: D992–999. 10.1093/nar/gkv1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao M, Kim P, Mitra R, Zhao J, Zhao Z (2016) TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res 44: D1023–1031. 10.1093/nar/gkv1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beerenwinkel N, Antal T, Dingli D, Traulsen A, Kinzler KW, et al. (2007) Genetic progression and the waiting time to cancer. PLoS Comput Biol 3: e225 10.1371/journal.pcbi.0030225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaminker JS, Zhang Y, Waugh A, Haverty PM, Peters B, et al. (2007) Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res 67: 465–473. 10.1158/0008-5472.CAN-06-1736 [DOI] [PubMed] [Google Scholar]
- 18.Stratton MR (2011) Exploring the genomes of cancer cells: progress and promise. Science 331: 1553–1558. 10.1126/science.1204040 [DOI] [PubMed] [Google Scholar]
- 19.Marx V (2013) Drilling into big cancer-genome data. Nat Methods 10: 293–297. 10.1038/nmeth.2410 [DOI] [PubMed] [Google Scholar]
- 20.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, et al. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, et al. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang J, Finney RP, Rowe W, Edmonson M, Yang SH, et al. (2007) Systematic analysis of genetic alterations in tumors using Cancer Genome WorkBench (CGWB). Genome Res 17: 1111–1117. 10.1101/gr.5963407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cline MS, Craft B, Swatloski T, Goldman M, Ma S, et al. (2013) Exploring TCGA Pan-Cancer data at the UCSC Cancer Genomics Browser. Sci Rep 3: 2652 10.1038/srep02652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, et al. (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218. 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., et al. (2013) Cancer genome landscapes. Science 339: 1546–1558. 10.1126/science.1235122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garraway LA, Lander ES (2013) Lessons from the cancer genome. Cell 153: 17–37. 10.1016/j.cell.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 27.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, et al. (2010) Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26: i237–245. 10.1093/bioinformatics/btq182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen QY, Jiao DM, Wu YQ, Wang L, Hu HZ, et al. (2013) Functional and pathway enrichment analysis for integrated regulatory network of high- and low-metastatic lung cancer. Mol Biosyst 9: 3080–3090. 10.1039/c3mb70288j [DOI] [PubMed] [Google Scholar]
- 29.Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, et al. (2015) Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 47: 106–114. 10.1038/ng.3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Torkamani A, Schork NJ (2009) Identification of rare cancer driver mutations by network reconstruction. Genome Res 19: 1570–1578. 10.1101/gr.092833.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu TJ, Schriml LM, Chen QR, Colbert M, Crichton DJ, et al. (2015) Generating a focused view of disease ontology cancer terms for pan-cancer data integration and analysis. Database (Oxford) 2015: bav032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu G, Feng X, Stein L (2010) A human functional protein interaction network and its application to cancer data analysis. Genome Biol 11: R53 10.1186/gb-2010-11-5-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kar G, Gursoy A, Keskin O (2009) Human cancer protein-protein interaction network: a structural perspective. PLoS Comput Biol 5: e1000601 10.1371/journal.pcbi.1000601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, et al. (2009) Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 69: 6660–6667. 10.1158/0008-5472.CAN-09-1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding J, Bashashati A, Roth A, Oloumi A, Tse K, et al. (2012) Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinformatics 28: 167–175. 10.1093/bioinformatics/btr629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Capriotti E, Altman RB (2011) A new disease-specific machine learning approach for the prediction of cancer-causing missense variants. Genomics 98: 310–317. 10.1016/j.ygeno.2011.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nehrt NL, Peterson TA, Park D, Kann MG (2012) Domain landscapes of somatic mutations in cancer. BMC Genomics 13 Suppl 4: S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang F, Petsalaki E, Rolland T, Hill DE, Vidal M, et al. (2015) Protein domain-level landscape of cancer-type-specific somatic mutations. PLoS Comput Biol 11: e1004147 10.1371/journal.pcbi.1004147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holm L, Sander C (1996) Mapping the protein universe. Science 273: 595–603. [DOI] [PubMed] [Google Scholar]
- 40.Murzin AG, Brenner SE, Hubbard T, Chothia C (1995) SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol 247: 536–540. 10.1006/jmbi.1995.0159 [DOI] [PubMed] [Google Scholar]
- 41.Diella F, Haslam N, Chica C, Budd A, Michael S, et al. (2008) Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front Biosci 13: 6580–6603. [DOI] [PubMed] [Google Scholar]
- 42.Zhong Q, Simonis N, Li QR, Charloteaux B, Heuze F, et al. (2009) Edgetic perturbation models of human inherited disorders. Mol Syst Biol 5: 321 10.1038/msb.2009.80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson TA, Nehrt NL, Park D, Kann MG (2012) Incorporating molecular and functional context into the analysis and prioritization of human variants associated with cancer. J Am Med Inform Assoc 19: 275–283. 10.1136/amiajnl-2011-000655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peterson TA, Adadey A, Santana-Cruz I, Sun Y, Winder A, et al. (2010) DMDM: domain mapping of disease mutations. Bioinformatics 26: 2458–2459. 10.1093/bioinformatics/btq447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson TA, Park D, Kann MG (2013) A protein domain-centric approach for the comparative analysis of human and yeast phenotypically relevant mutations. BMC Genomics 14 Suppl 3: S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amberger J, Bocchini C, Hamosh A (2011) A new face and new challenges for Online Mendelian Inheritance in Man (OMIM(R)). Hum Mutat 32: 564–567. 10.1002/humu.21466 [DOI] [PubMed] [Google Scholar]
- 47.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, et al. (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res 31: 365–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, et al. (2013) IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods 10: 1081–1082. 10.1038/nmeth.2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4: 1073–1081. 10.1038/nprot.2009.86 [DOI] [PubMed] [Google Scholar]
- 50.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A method and server for predicting damaging missense mutations. Nat Methods 7: 248–249. 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, et al. (2010) Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070. 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39: e118 10.1093/nar/gkr407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Collins FS, Barker AD (2007) Mapping the cancer genome. Pinpointing the genes involved in cancer will help chart a new course across the complex landscape of human malignancies. Sci Am 296: 50–57. [PubMed] [Google Scholar]
- 54.Pruitt KD, Tatusova T, Brown GR, Maglott DR (2012) NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res 40: D130–135. 10.1093/nar/gkr1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.(2013) UniProtKB/Swiss-Prot protein knowledgebase release 2013_05 statistics.
- 56.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, et al. (2010) The NCBI BioSystems database. Nucleic Acids Res 38: D492–496. 10.1093/nar/gkp858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, et al. (2013) CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41: D348–352. 10.1093/nar/gks1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RD, Mistry J, Tate J, Coggill P, Heger A, et al. (2010) The Pfam protein families database. Nucleic Acids Res 38: D211–222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eddy SR (1996) Hidden markov models. Current opinion in structural biology 6: 361–365. [DOI] [PubMed] [Google Scholar]
- 60.Yue P, Forrest WF, Kaminker JS, Lohr S, Zhang Z, et al. (2010) Inferring the functional effects of mutation through clusters of mutations in homologous proteins. Hum Mutat 31: 264–271. 10.1002/humu.21194 [DOI] [PubMed] [Google Scholar]
- 61.Gauran II, Park J, Lim J, Park D, Zylstra J, et al. (2016) Empirical Null Estimation using Discrete Mixture Distributions and its Application to Protein Domain Data. ArXiv e-prints. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Supek F, Minana B, Valcarcel J, Gabaldon T, Lehner B (2014) Synonymous mutations frequently act as driver mutations in human cancers. Cell 156: 1324–1335. 10.1016/j.cell.2014.01.051 [DOI] [PubMed] [Google Scholar]
- 63.Gotea V, Gartner JJ, Qutob N, Elnitski L, Samuels Y (2015) The functional relevance of somatic synonymous mutations in melanoma and other cancers. Pigment Cell Melanoma Res 28: 673–684. 10.1111/pcmr.12413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gartner JJ, Parker SC, Prickett TD, Dutton-Regester K, Stitzel ML, et al. (2013) Whole-genome sequencing identifies a recurrent functional synonymous mutation in melanoma. Proc Natl Acad Sci U S A 110: 13481–13486. 10.1073/pnas.1304227110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pei J, Grishin NV (2001) AL2CO: calculation of positional conservation in a protein sequence alignment. Bioinformatics 17: 700–712. [DOI] [PubMed] [Google Scholar]
- 66.(2015) Gene Ontology Consortium: going forward. Nucleic Acids Res 43: D1049–1056. 10.1093/nar/gku1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Glazko GV, Babenko VN, Koonin EV, Rogozin IB (2006) Mutational hotspots in the TP53 gene and, possibly, other tumor suppressors evolve by positive selection. Biol Direct 1: 4 10.1186/1745-6150-1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, et al. (2000) APC mutations in sporadic colorectal tumors: A mutational "hotspot" and interdependence of the "two hits". Proc Natl Acad Sci U S A 97: 3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, et al. (2016) Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol 34: 155–163. 10.1038/nbt.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hall A (2012) Rho family gtpases. Biochemical Society Transactions 40: 1378–1382. 10.1042/BST20120103 [DOI] [PubMed] [Google Scholar]
- 71.Alan JK, Lundquist EA (2013) Mutationally activated Rho GTPases in cancer. Small GTPases 4: 159–163. 10.4161/sgtp.26530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Okegawa T, Pong RC, Li Y, Hsieh JT (2004) The role of cell adhesion molecule in cancer progression and its application in cancer therapy. Acta Biochim Pol 51: 445–457. [PubMed] [Google Scholar]
- 73.Farahani E, Patra HK, Jangamreddy JR, Rashedi I, Kawalec M, et al. (2014) Cell adhesion molecules and their relation to (cancer) cell stemness. Carcinogenesis 35: 747–759. 10.1093/carcin/bgu045 [DOI] [PubMed] [Google Scholar]
- 74.Knights AJ, Funnell AP, Crossley M, Pearson RC (2012) Holding Tight: Cell Junctions and Cancer Spread. Trends Cancer Res 8: 61–69. [PMC free article] [PubMed] [Google Scholar]
- 75.Bendas G, Borsig L (2012) Cancer cell adhesion and metastasis: selectins, integrins, and the inhibitory potential of heparins. Int J Cell Biol 2012: 676731 10.1155/2012/676731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin TA, Mason MD, Jiang WG (2011) Tight junctions in cancer metastasis. Front Biosci (Landmark Ed) 16: 898–936. [DOI] [PubMed] [Google Scholar]
- 77.Cairns RA, Harris IS, Mak TW (2011) Regulation of cancer cell metabolism. Nat Rev Cancer 11: 85–95. 10.1038/nrc2981 [DOI] [PubMed] [Google Scholar]
- 78.Seyfried TN, Flores RE, Poff AM, D'Agostino DP (2014) Cancer as a metabolic disease: implications for novel therapeutics. Carcinogenesis 35: 515–527. 10.1093/carcin/bgt480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Phan LM, Yeung SC, Lee MH (2014) Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med 11: 1–19. 10.7497/j.issn.2095-3941.2014.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Denys H, Braems G, Lambein K, Pauwels P, Hendrix A, et al. (2009) The extracellular matrix regulates cancer progression and therapy response: implications for prognosis and treatment. Curr Pharm Des 15: 1373–1384. [DOI] [PubMed] [Google Scholar]
- 81.Lu P, Weaver VM, Werb Z (2012) The extracellular matrix: a dynamic niche in cancer progression. J Cell Biol 196: 395–406. 10.1083/jcb.201102147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox TR, Erler JT (2011) Remodeling and homeostasis of the extracellular matrix: implications for fibrotic diseases and cancer. Dis Model Mech 4: 165–178. 10.1242/dmm.004077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oskarsson T (2013) Extracellular matrix components in breast cancer progression and metastasis. Breast 22 Suppl 2: S66–72. [DOI] [PubMed] [Google Scholar]
- 84.Minciacchi VR, Freeman MR, Di Vizio D (2015) Extracellular vesicles in cancer: exosomes, microvesicles and the emerging role of large oncosomes. Semin Cell Dev Biol 40: 41–51. 10.1016/j.semcdb.2015.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Green TM, Alpaugh ML, Barsky SH, Rappa G, Lorico A (2015) Breast Cancer-Derived Extracellular Vesicles: Characterization and Contribution to the Metastatic Phenotype. Biomed Res Int 2015: 634865 10.1155/2015/634865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ohta T, Fukuda M (2004) Ubiquitin and breast cancer. Oncogene 23: 2079–2088. 10.1038/sj.onc.1207371 [DOI] [PubMed] [Google Scholar]
- 87.Mani A, Gelmann EP (2005) The ubiquitin-proteasome pathway and its role in cancer. J Clin Oncol 23: 4776–4789. 10.1200/JCO.2005.05.081 [DOI] [PubMed] [Google Scholar]
- 88.Yerlikaya A, Yontem M (2013) The significance of ubiquitin proteasome pathway in cancer development. Recent Pat Anticancer Drug Discov 8: 298–309. [DOI] [PubMed] [Google Scholar]
- 89.Duffy MJ (1992) The role of proteolytic enzymes in cancer invasion and metastasis. Clin Exp Metastasis 10: 145–155. [DOI] [PubMed] [Google Scholar]
- 90.Wolf K, Friedl P (2005) Functional imaging of pericellular proteolysis in cancer cell invasion. Biochimie 87: 315–320. 10.1016/j.biochi.2004.10.016 [DOI] [PubMed] [Google Scholar]
- 91.Sevenich L, Joyce JA (2014) Pericellular proteolysis in cancer. Genes Dev 28: 2331–2347. 10.1101/gad.250647.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Reznik E, Sander C (2015) Extensive decoupling of metabolic genes in cancer. PLoS Comput Biol 11: e1004176 10.1371/journal.pcbi.1004176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Furuta E, Okuda H, Kobayashi A, Watabe K (2010) Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim Biophys Acta 1805: 141–152. 10.1016/j.bbcan.2010.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hall A (2009) The cytoskeleton and cancer. Cancer Metastasis Rev 28: 5–14. 10.1007/s10555-008-9166-3 [DOI] [PubMed] [Google Scholar]
- 95.Yamaguchi H, Condeelis J (2007) Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim Biophys Acta 1773: 642–652. 10.1016/j.bbamcr.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fife CM, McCarroll JA, Kavallaris M (2014) Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol 171: 5507–5523. 10.1111/bph.12704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rowinsky EK (2003) Signal events: Cell signal transduction and its inhibition in cancer. Oncologist 8 Suppl 3: 5–17. [DOI] [PubMed] [Google Scholar]
- 98.Sever R, Brugge JS (2015) Signal transduction in cancer. Cold Spring Harb Perspect Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kampen KR (2011) Membrane proteins: the key players of a cancer cell. J Membr Biol 242: 69–74. 10.1007/s00232-011-9381-7 [DOI] [PubMed] [Google Scholar]
- 100.Leth-Larsen R, Lund R, Hansen HV, Laenkholm AV, Tarin D, et al. (2009) Metastasis-related plasma membrane proteins of human breast cancer cells identified by comparative quantitative mass spectrometry. Mol Cell Proteomics 8: 1436–1449. 10.1074/mcp.M800061-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Neuhaus EM, Zhang W, Gelis L, Deng Y, Noldus J, et al. (2009) Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J Biol Chem 284: 16218–16225. 10.1074/jbc.M109.012096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanz G, Leray I, Dewaele A, Sobilo J, Lerondel S, et al. (2014) Promotion of cancer cell invasiveness and metastasis emergence caused by olfactory receptor stimulation. PLoS One 9: e85110 10.1371/journal.pone.0085110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morita R, Hirohashi Y, Torigoe T, Inoda S, Takahashi A, et al. (2016) Olfactory receptor family receptor, family 7, subfamily C, member 1 is a novel marker of colon cancer-initiating cells and is a potent target of immunotherapy. Clin Cancer Res. [DOI] [PubMed] [Google Scholar]
- 104.Weng J, Ma W, Mitchell D, Zhang J, Liu M (2005) Regulation of human prostate-specific G-protein coupled receptor, PSGR, by two distinct promoters and growth factors. J Cell Biochem 96: 1034–1048. 10.1002/jcb.20600 [DOI] [PubMed] [Google Scholar]
- 105.Cardillo MR, Di Silverio F (2006) Prostate—specific G protein couple receptor genes and STAG1/PMEPA1 in peripheral blood from patients with prostatic cancer. Int J Immunopathol Pharmacol 19: 871–878. 10.1177/039463200601900416 [DOI] [PubMed] [Google Scholar]
- 106.Li J, Mahajan A, Tsai MD (2006) Ankyrin repeat: a unique motif mediating protein-protein interactions. Biochemistry 45: 15168–15178. 10.1021/bi062188q [DOI] [PubMed] [Google Scholar]
- 107.Wang Y, McKay JD, Rafnar T, Wang Z, Timofeeva MN, et al. (2014) Rare variants of large effect in BRCA2 and CHEK2 affect risk of lung cancer. Nat Genet 46: 736–741. 10.1038/ng.3002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bodmer W, Tomlinson I (2010) Rare genetic variants and the risk of cancer. Curr Opin Genet Dev 20: 262–267. 10.1016/j.gde.2010.04.016 [DOI] [PubMed] [Google Scholar]
- 109.Kim WK, Bang MH, Kim ES, Kang NE, Jung KC, et al. (2005) Quercetin decreases the expression of ErbB2 and ErbB3 proteins in HT-29 human colon cancer cells. J Nutr Biochem 16: 155–162. 10.1016/j.jnutbio.2004.10.010 [DOI] [PubMed] [Google Scholar]
- 110.Yonesaka K, Zejnullahu K, Okamoto I, Satoh T, Cappuzzo F, et al. (2011) Activation of ERBB2 signaling causes resistance to the EGFR-directed therapeutic antibody cetuximab. Sci Transl Med 3: 99ra86 10.1126/scitranslmed.3002442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Markman B, Javier Ramos F, Capdevila J, Tabernero J (2010) EGFR and KRAS in colorectal cancer. Adv Clin Chem 51: 71–119. [DOI] [PubMed] [Google Scholar]
- 112.Spano JP, Fagard R, Soria JC, Rixe O, Khayat D, et al. (2005) Epidermal growth factor receptor signaling in colorectal cancer: preclinical data and therapeutic perspectives. Ann Oncol 16: 189–194. 10.1093/annonc/mdi057 [DOI] [PubMed] [Google Scholar]
- 113.Gavert N, Shvab A, Sheffer M, Ben-Shmuel A, Haase G, et al. (2013) c-Kit is suppressed in human colon cancer tissue and contributes to L1-mediated metastasis. Cancer Res 73: 5754–5763. 10.1158/0008-5472.CAN-13-0576 [DOI] [PubMed] [Google Scholar]
- 114.Akintola-Ogunremi O, Pfeifer JD, Tan BR, Yan Y, Zhu X, et al. (2003) Analysis of protein expression and gene mutation of c-kit in colorectal neuroendocrine carcinomas. Am J Surg Pathol 27: 1551–1558. [DOI] [PubMed] [Google Scholar]
- 115.Kalady MF, Dejulius KL, Sanchez JA, Jarrar A, Liu X, et al. (2012) BRAF mutations in colorectal cancer are associated with distinct clinical characteristics and worse prognosis. Dis Colon Rectum 55: 128–133. 10.1097/DCR.0b013e31823c08b3 [DOI] [PubMed] [Google Scholar]
- 116.Barras D (2015) BRAF Mutation in Colorectal Cancer: An Update. Biomark Cancer 7: 9–12. 10.4137/BIC.S25248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pietrantonio F, Petrelli F, Coinu A, Di Bartolomeo M, Borgonovo K, et al. (2015) Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer 51: 587–594. 10.1016/j.ejca.2015.01.054 [DOI] [PubMed] [Google Scholar]
- 118.Luo Y, Tsuchiya KD, Il Park D, Fausel R, Kanngurn S, et al. (2013) RET is a potential tumor suppressor gene in colorectal cancer. Oncogene 32: 2037–2047. 10.1038/onc.2012.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, et al. (2012) Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med 18: 382–384. 10.1038/nm.2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Grady WM, Willis JE, Trobridge P, Romero-Gallo J, Munoz N, et al. (2006) Proliferation and Cdk4 expression in microsatellite unstable colon cancers with TGFBR2 mutations. Int J Cancer 118: 600–608. 10.1002/ijc.21399 [DOI] [PubMed] [Google Scholar]
- 121.Ye YJ, Zhu XG, Wang S, Wang YC, Sang JL (2006) [Antisense to CDK4 inhibits the growth of human colon cancer cells HT29]. Zhonghua Yi Xue Za Zhi 86: 846–849. [PubMed] [Google Scholar]
- 122.Zhao P, Hu YC, Talbot IC (2003) Expressing patterns of p16 and CDK4 correlated to prognosis in colorectal carcinoma. World J Gastroenterol 9: 2202–2206. 10.3748/wjg.v9.i10.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Aisner DL, Nguyen TT, Paskulin DD, Le AT, Haney J, et al. (2014) ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res 12: 111–118. 10.1158/1541-7786.MCR-13-0479-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bavi P, Jehan Z, Bu R, Prabhakaran S, Al-Sanea N, et al. (2013) ALK gene amplification is associated with poor prognosis in colorectal carcinoma. Br J Cancer 109: 2735–2743. 10.1038/bjc.2013.641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Alese OB, El-Rayes BF, Sica G, Zhang G, Alexis D, et al. (2015) Anaplastic lymphoma kinase (ALK) gene alteration in signet ring cell carcinoma of the gastrointestinal tract. Ther Adv Med Oncol 7: 56–62. 10.1177/1758834014567117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Slattery ML, Lundgreen A, Wolff RK (2013) Dietary influence on MAPK-signaling pathways and risk of colon and rectal cancer. Nutr Cancer 65: 729–738. 10.1080/01635581.2013.795599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ohmori M, Shirasawa S, Furuse M, Okumura K, Sasazuki T (1997) Activated Ki-ras enhances sensitivity of ceramide-induced apoptosis without c-Jun NH2-terminal kinase/stress-activated protein kinase or extracellular signal-regulated kinase activation in human colon cancer cells. Cancer Res 57: 4714–4717. [PubMed] [Google Scholar]
- 128.Ahmed N, Oliva K, Wang Y, Quinn M, Rice G (2003) Downregulation of urokinase plasminogen activator receptor expression inhibits Erk signalling with concomitant suppression of invasiveness due to loss of uPAR-beta1 integrin complex in colon cancer cells. Br J Cancer 89: 374–384. 10.1038/sj.bjc.6601098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sugimura M, Kobayashi K, Sagae S, Nishioka Y, Ishioka S, et al. (2000) Mutation of the SRC gene in endometrial carcinoma. Jpn J Cancer Res 91: 395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lau GM, Yu GL, Gelman IH, Gutowski A, Hangauer D, et al. (2009) Expression of Src and FAK in hepatocellular carcinoma and the effect of Src inhibitors on hepatocellular carcinoma in vitro. Dig Dis Sci 54: 1465–1474. 10.1007/s10620-008-0519-0 [DOI] [PubMed] [Google Scholar]
- 131.Chen ML, Chai CY, Yeh KT, Wang SN, Tsai CJ, et al. (2011) Crosstalk between activated and inactivated c-Src in hepatocellular carcinoma. Dis Markers 30: 325–333. 10.3233/DMA-2011-0792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lee SH, Lee JW, Soung YH, Kim HS, Park WS, et al. (2003) BRAF and KRAS mutations in stomach cancer. Oncogene 22: 6942–6945. 10.1038/sj.onc.1206749 [DOI] [PubMed] [Google Scholar]
- 133.Lee SH, Ahn BK, Baek SU, Chang HK (2013) BRAF mutation in multiple primary cancer with colorectal cancer and stomach cancer. Gastroenterol Rep (Oxf) 1: 70–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Balschun K, Haag J, Wenke AK, von Schonfels W, Schwarz NT, et al. (2011) KRAS, NRAS, PIK3CA exon 20, and BRAF genotypes in synchronous and metachronous primary colorectal cancers diagnostic and therapeutic implications. J Mol Diagn 13: 436–445. 10.1016/j.jmoldx.2011.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dahiya S, Emnett RJ, Haydon DH, Leonard JR, Phillips JJ, et al. (2014) BRAF-V600E mutation in pediatric and adult glioblastoma. Neuro Oncol 16: 318–319. 10.1093/neuonc/not146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Takahashi Y, Akahane T, Sawada T, Ikeda H, Tempaku A, et al. (2015) Adult classical glioblastoma with a BRAF V600E mutation. World J Surg Oncol 13: 100 10.1186/s12957-015-0521-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Suzuki Y, Takahashi-Fujigasaki J, Akasaki Y, Matsushima S, Mori R, et al. (2015) BRAF V600E-mutated diffuse glioma in an adult patient: a case report and review. Brain Tumor Pathol. [DOI] [PubMed] [Google Scholar]
- 138.He M, Breese V, Hang S, Zhang C, Xiong J, et al. (2013) BRAF V600E Mutations in Endometrial Adenocarcinoma. Diagn Mol Pathol 22: 35–40. 10.1097/PDM.0b013e31826c7fe0 [DOI] [PubMed] [Google Scholar]
- 139.Feng YZ, Shiozawa T, Miyamoto T, Kashima H, Kurai M, et al. (2005) BRAF mutation in endometrial carcinoma and hyperplasia: correlation with KRAS and p53 mutations and mismatch repair protein expression. Clin Cancer Res 11: 6133–6138. 10.1158/1078-0432.CCR-04-2670 [DOI] [PubMed] [Google Scholar]
- 140.Gao M, Liang XJ, Zhang ZS, Ma W, Chang ZW, et al. (2013) Relationship between expression of EGFR in gastric cancer tissue and clinicopathological features. Asian Pac J Trop Med 6: 260–264. 10.1016/S1995-7645(13)60054-1 [DOI] [PubMed] [Google Scholar]
- 141.Liu Z, Liu L, Li M, Wang Z, Feng L, et al. (2011) Epidermal growth factor receptor mutation in gastric cancer. Pathology 43: 234–238. 10.1097/PAT.0b013e328344e61b [DOI] [PubMed] [Google Scholar]
- 142.Gaffney DC, Soyer HP, Simpson F (2014) The epidermal growth factor receptor in squamous cell carcinoma: An emerging drug target. Australas J Dermatol 55: 24–34. 10.1111/ajd.12025 [DOI] [PubMed] [Google Scholar]
- 143.Boone B, Jacobs K, Ferdinande L, Taildeman J, Lambert J, et al. (2011) EGFR in melanoma: clinical significance and potential therapeutic target. J Cutan Pathol 38: 492–502. 10.1111/j.1600-0560.2011.01673.x [DOI] [PubMed] [Google Scholar]
- 144.Gross A, Niemetz-Rahn A, Nonnenmacher A, Tucholski J, Keilholz U, et al. (2015) Expression and activity of EGFR in human cutaneous melanoma cell lines and influence of vemurafenib on the EGFR pathway. Target Oncol 10: 77–84. 10.1007/s11523-014-0318-9 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency distribution of the number of times pfam oncodomain families form a hotspot in 20 different cancer types.
(TIF)
Visual representation and hierarchical clustering of oncodomain hotspots on genes that were significant in CHASM or MutSigCV. For each cell, the ratio of patients with somatic variants in a hotspot to patients with a somatic variant in the domain region is quantified. Each cell is color-coded if the gene had any somatic variants of that cancer type in an oncodomain hotspot (blue), if it was significant in CHASM/MutSigCV (green), or both (purple). Only the top ten genes based on the gene name’s co-occurrence with the “cancer” MeSH term are shown. Here, cancer types are grouped via hierarchical clustering to show similar mutational patterns. Enumerated in each cell are the proportion of patients with a somatic variant in an oncodomain hotspot (numerator) compared to the number of patients that had a somatic variant anywhere in the protein domain region (denominator).
(TIF)
Bootstrap analysis was performed to count the number of Pfam oncodomains and oncodomain hotspots with only 75% or 50% of the available patients or available exonic somatic variants. The bootstrapping process was repeated 100 times for each cancer type, bootstrap percentage, and local false discovery rate cutoffs.
(DOCX)
Enrichment of the Biological Process and Molecular Function Gene Ontology ontologies for genes with at least one somatic variant in an oncodomain hotspot for any cancer type.
(DOCX)
Top twenty enriched Gene Ontology terms with Pfam oncodomains from the pfam2go annotations using Fisher’s exact test with Bonferroni correction.
(DOCX)
(XLSX)
(ZIP)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.