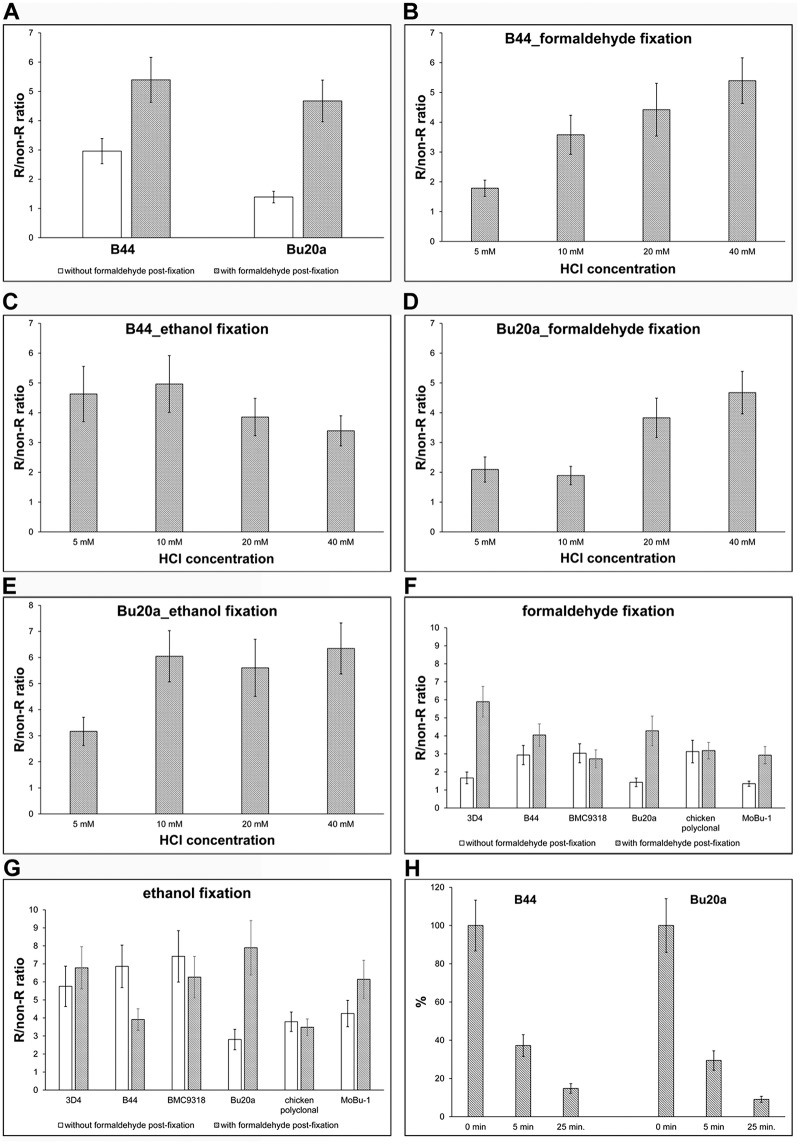
Fig 1. Optimising the protocol for BrdU detection.
(A) HeLa cells were incubated with BrdU for 30 min, fixed with formaldehyde and BrdU was detected in DNA using 40 mM HCl. BrdU was detected either by the B44 or Bu20a anti-BrdU antibody with exonuclease III. The effect of formaldehyde post-fixation on the signal is shown as well. The data are presented as the mean ± SD. (B-E) A comparison of various HCl concentrations on BrdU signal in HeLa cells labelled for 30 min with BrdU and fixed either with formaldehyde (B, D) or ethanol (C, E). The incorporated BrdU was detected using either B44 (B, C) or Bu20a (D, E) antibody clone with exonuclease III. The data are presented as the mean ± SD. (F, G) A comparison of five monoclonal anti-BrdU antibody clones and one polyclonal antibody is shown. The HeLa cells were labelled with BrdU for 30 min and fixed either with formaldehyde (F) or ethanol (G). The impact of the post-fixation step is shown as well. The data are presented as the mean ± SD. (H) The effect of the length of the washing step on the BrdU-derived signal. The HeLa cells were labelled with BrdU for 30 min and fixed with formaldehyde. After incubation with the primary antibody, the cells were washed for 5 s (0 min) or 5 or 25 min in 1× PBS and then post-fixed with formaldehyde. The data are normalised to the % of the average signal in samples washed for 5 s in 1× PBS and then immediately post-fixed with formaldehyde. The data are presented as the mean ± SD.