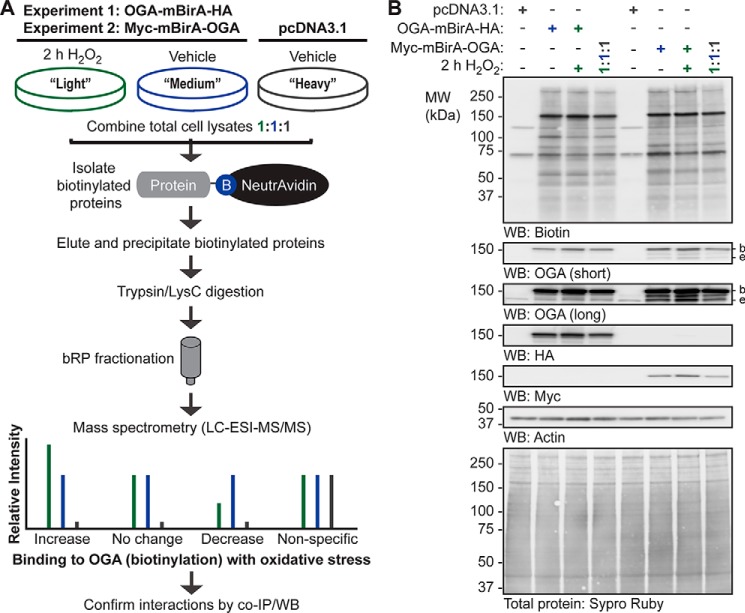
Figure 5.
SILAC-BioID-MS/MS strategy used to identify the basal and oxidative stress-dependent interactome of OGA. A, U2OS cells were labeled with light, medium, or heavy isotopes of arginine and lysine for six generations. In experiment 1, cells were transfected with pcDNA3.1 (heavy) or OGA-mBirA-HA (light, medium), treated with biotin (25 μm, 16 h), and treated with vehicle (medium, heavy) or H2O2 (light; 2.5 mm, 2 h, n = 1). In experiment 2, Myc-mBirA-OGA was transfected in replacement of OGA-mBirA-HA. For each experiment, proteins were extracted in denaturing TCL buffer and combined in equal amounts. The biotinylated proteins were isolated on NeutrAvidin-agarose in denaturing conditions, eluted in 2% (w/v) SDS (95 °C), and precipitated with acetone. Peptides were generated by trypsin and LysC digestion, separated by basic reversed phase (bRP) fractionation, and identified by mass spectrometry (LC-ESI-MS/MS). Subsequently, protein-protein interactions were validated by co- IP and Western blotting. B, for the SILAC experiments (n = 1, each), equal amounts of protein (10 μg; denaturing TCL lysis) were separated by SDS-PAGE, and the following were detected by Western blotting: biotin, OGA, HA, Myc, and actin. Protein load was assessed by total protein stain (Sypro Ruby) and by Western blotting (actin). Migration of endogenous OGA (e), mBirA-tagged OGA (b), and the molecular mass (MW) markers are indicated.