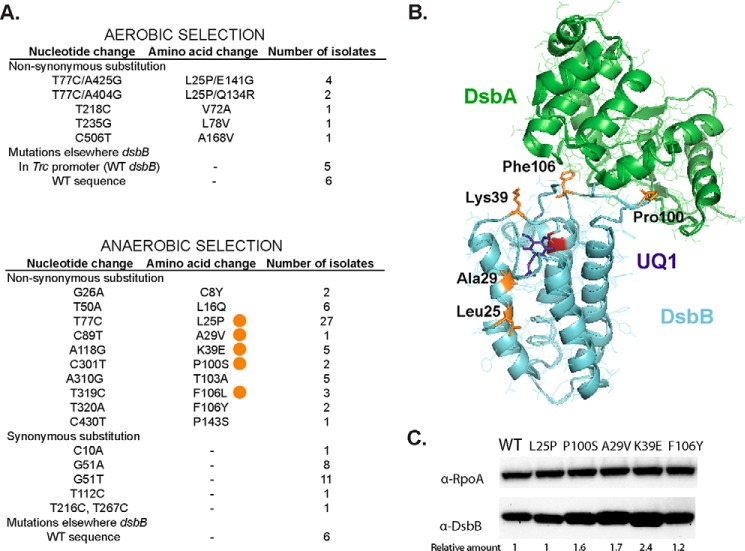
Figure 2.
Compound 12-resistant mutations using two different selections. A, nucleotide changes found in dsbB after selection of LptD4213 growth on aerobic minimal media in the presence of 10 μm compound 12 (top). Nucleotide changes found in dsbB after selection on anaerobic minimal media in the presence of 2 μm compound 12 (bottom). The mutations studied in this work are indicated with an orange circle. B, location of the mutations (orange) in the structure of the DsbA-DsbB complex. DsbA is shown in green, DsbB in cyan, Cys-44 in red, and ubiquinone-1 (UQ1) in purple. PyMOL was used to visualize the structure (2ZUP) of the crystallized complex when Cys-30 of DsbA is forming a disulfide bond with Cys-104 of DsbB and Cys-41 and Cys-44 of DsbB are disulfide bonded. C, α-DsbB immunoblot analysis of strains carrying dsbB at λ att site under the control of trc204 promoter (CL591 to CL596 strains). Cells were grown for 4 h in M63 minimal medium with 0.2% glucose and 1 mm IPTG to induce expression of DsbB. Cells were TCA-precipitated, and protein pellets were resuspended in 100 mm Tris, 1% SDS buffer. β-Mercaptoethanol was used to reduce the proteins. 10 μg of total protein samples were loaded in 12% acrylamide gel. α-RpoA was used as a loading control. The relative amount was calculated using arbitrary levels given by Image Lab 5.2 software.