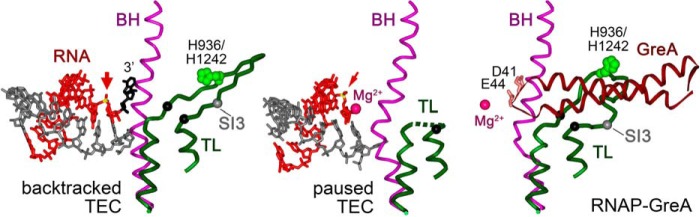
Figure 1.
Structures of backtracked TEC (PDB code 4WQS, left), elemental paused TEC (PDB code 4GZY, center), and GreA-bound TthRNAP (PDB code 4WQT, right) (9, 28). The TL, BH, and GreA are dark green, violet, and dark red, respectively. The His-936/His-1242 residue in the TL is light green, The Cα atoms at the borders of the TL deletions analyzed in our work (Δ931–1137→3Gly, Δ1254–1272→3Gly, and Δ1237–1255→3Ala in E. coli, D. radiodurans, and T. aquaticus RNAPs, respectively) are shown with black spheres. The position of the SI3 insertion in EcoRNAP is shown as a gray sphere. RNA is red, the 3′-nucleotide in the backtracked TEC is black, the sites of RNA cleavage are indicated with red arrows, and the attacked phosphorus atom is yellow. Mg2+ ions in the RNAP active site are shown with carmine spheres (note that no Me2+ atoms were resolved in the backtracked TEC, and only the first ion is visible in the paused and Gre-bound RNAPs). Acidic residues at the tip of the GreA NTD (Asp-41, Glu-44) are pink. In the paused TEC structure, the central part of the TL is disordered, and the ultimate RNA phosphodiester bond is shifted upstream of the active center (28).