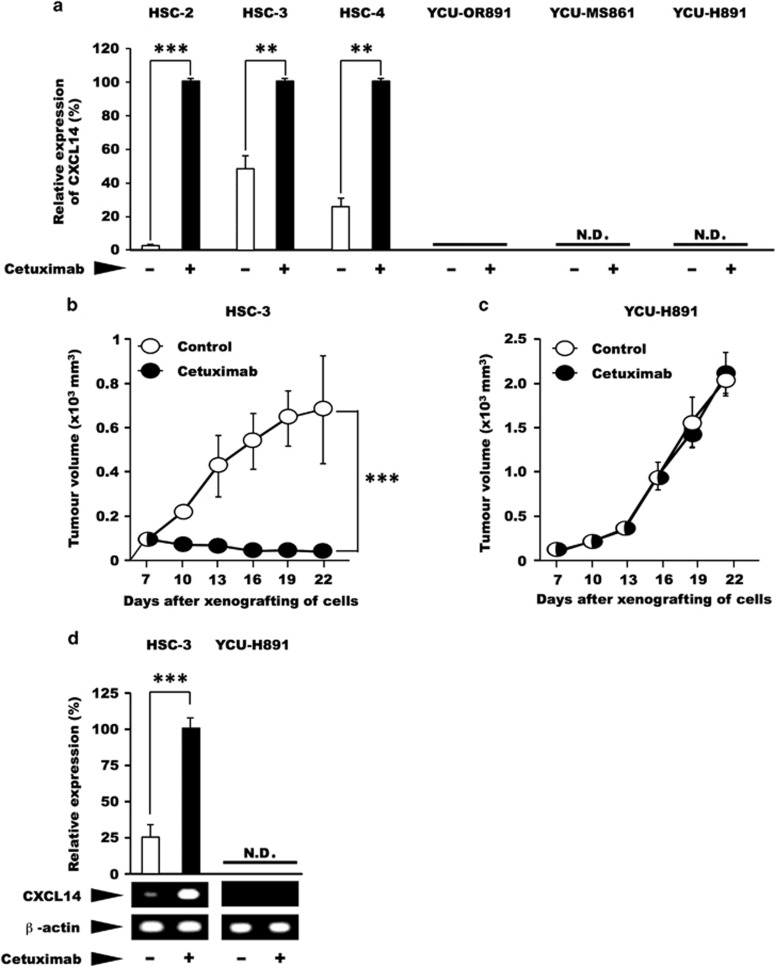
Figure 1.
Effects of cetuximab on the expression level of CXCL14 mRNA and tumour volume. (a) In an in vitro experiment, oral floor carcinoma-derived HSC-2 cells, tongue carcinoma-derived HSC-3 and HSC-4 cells, obtained from the Japanese Collection of Research Bioresources Cell Bank, as well as oral floor carcinoma-derived YCU-OR891 cells, maxillary sinus carcinoma-derived YCU-MS861 cells and hypopharynx carcinoma-derived YCU-H891 cells, which were established in our laboratory in a previous study,39 were cultured in Dulbecco's Modified Eagle's medium (DMEM) (Sigma-Aldrich, St Louis, MO, USA) containing 50 μg/ml gentamicin sulphate (Wako Pure Chemical Industry, Osaka, Japan) and 10% fetal bovine serum (Wako or Thermo Fisher Scientific, Yokohama, Japan) at 37 °C under 95% air and 5% CO2 until the cells reached the pre-confluent stage. The cells (1 × 105 per well) were then inoculated into three wells of six-well plates (Corning, Tokyo, Japan). After 2 days in culture, the cells were treated with cetuximab (10 μg/ml Bristol-Myers Squibb Company, New York City, NY, USA) or control for 24 h. Total RNA was subsequently extracted and purified using TRIzol (Thermo Fisher Scientific). Total RNA (1 μg) was reverse transcribed to complementary DNA (cDNA) using a PrimeScript RT Reagent kit with gDNA Eraser (Perfect Real Time: Takara, Kusatsu, Japan). Reactions for reverse transcription proceeded according to the manufacturer's instructions: 42 °C (gDNA removal) for 2 min, 37 °C (reverse transcription) for 15 min and 85 °C (inactivation of the reverse transcriptase) for 5 s. All quantitative PCR (qPCR) experiments were performed using an Applied Biosystems StepOnePlus Real-Time PCR Systems (Applied Biosystems, Tokyo, Japan). All amplifications were performed with Power SYBR Green PCR Master Mix (Life Technologies, Warrington, UK). Primer sets were as follows: for human CXCL14, 5′-AAGCCAAAGTACCCGCACTG-3′ (forward) and 5′-GACCTCGGTACCTGGACACG-3′ (reverse), which yielded a 73-bp product; and for human β-actin, 5′-GTGAAGGTGACAGCAGTCGGTT-3′ (forward) and 5′-GAAGTGGGGTGGCTTTTAGGAT-3′ (reverse), which yielded a 157-bp product. The thermal cycling conditions included an initial denaturation step at 95 °C for 10 min, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. Melting curve analysis of every qPCR was conducted after each cycle. Specific amplification was confirmed by checking the melting curves and melting temperatures of the qPCR products. Experiments were performed in triplicate, and the values were normalized to β-actin. (b, c) For the in vivo experiments, we subcutaneously inoculated BALB/c nude mice (female, 5-week old, Clea Japan Inc., Tokyo, Japan) on the dorsal side with 1 × 107 HSC-3 or YCU-H891 cells (12 mice per group). Seven days after cell inoculation (at a tumour size of ~100 mm3), we intraperitoneally administered cetuximab (10 mg/kg) or Dulbecco's phosphate-buffered saline (DPBS, Wako) at random to the animals three times per week and measured the tumour size once every 3 days for the HSC-3 cells (b) and YCU-H891 cells (c). Tumour volumes were measured by a person different from the one who injected cetuximab or DPBS once every 3 days and were calculated using the formula, (a × b × b)/2, where ‘a' is the long diameter and ‘b' is its short diameter of the tumours. (d) To determine the expression levels of CXCL14 mRNA in vivo, we removed the tumours 22 days after inoculation, isolated the total RNA and measured the expression level of CXCL14 mRNA in the HSC-3 and YCU-H891 tumours. CXCL14 cDNA was synthesized by performing the reverse transcription–PCR (RT–PCR) with SuperScript II reverse transcriptase (Invitrogen, Thermo Fisher Scientific K.K. Yokohama, Japan) and Ex Taq DNA polymerase (Takara, Otsu, Japan). Brilliant SYBR Green qPCR Master Mix was obtained from Stratagene (La Jolla, CA, USA). The following primers were used for RT–PCR: primers for human CXCL14, 5′-AAT GAA GCC AAA GTA CCC GC-3′ (forward) and 5′-AGT CCT TTG CAC AAG TCT CC-3′ (reverse; PCR product size, 230 bp); and primers for β-actin, 5′-AAA GAC CTG TAC GCC AAC AC-3′ (forward) and 5′-CTC GTC ATA CTC CTG CTT GC-3′ (reverse; PCR product size, 222 bp). The PCR cycling conditions included denaturation at 94 °C for 30 s, annealing at 58 °C for 30 s and elongation at 72 °C for 30 s. The PCR products were separated on 2% agarose gel by electrophoresis and were visualized with ethidium bromide dye.45 β-Actin cDNA was used as an internal standard and for normalisation. qPCR and/or densitometry were employed for quantitative comparison of the expression levels of CXCL14 mRNA between the two groups. These experiments were repeated twice. The in vivo experiments were performed in accordance with the local guidelines for the welfare of experimental animals and with the approval of the Ethics Committee on Animal Research of Kanagawa Dental University. The animals were housed in temperature-controlled rooms and received water and food ad libitum. We followed the guidelines for animal research of the International Association for the Study of Pain Committee for Research and Ethical Issues. In these experiments, Student's t-test was used to evaluate statistically significant differences between any two groups. N.D.; not detected. The values are expressed as the mean±s.d. (n=6). Half-white, half-black circles represent overlapping points. A P-value<0.05 was considered statistically significant. ***P<0.001 and **P<0.01. For some of the data points, the s.d. values were smaller than the size of the symbols used.