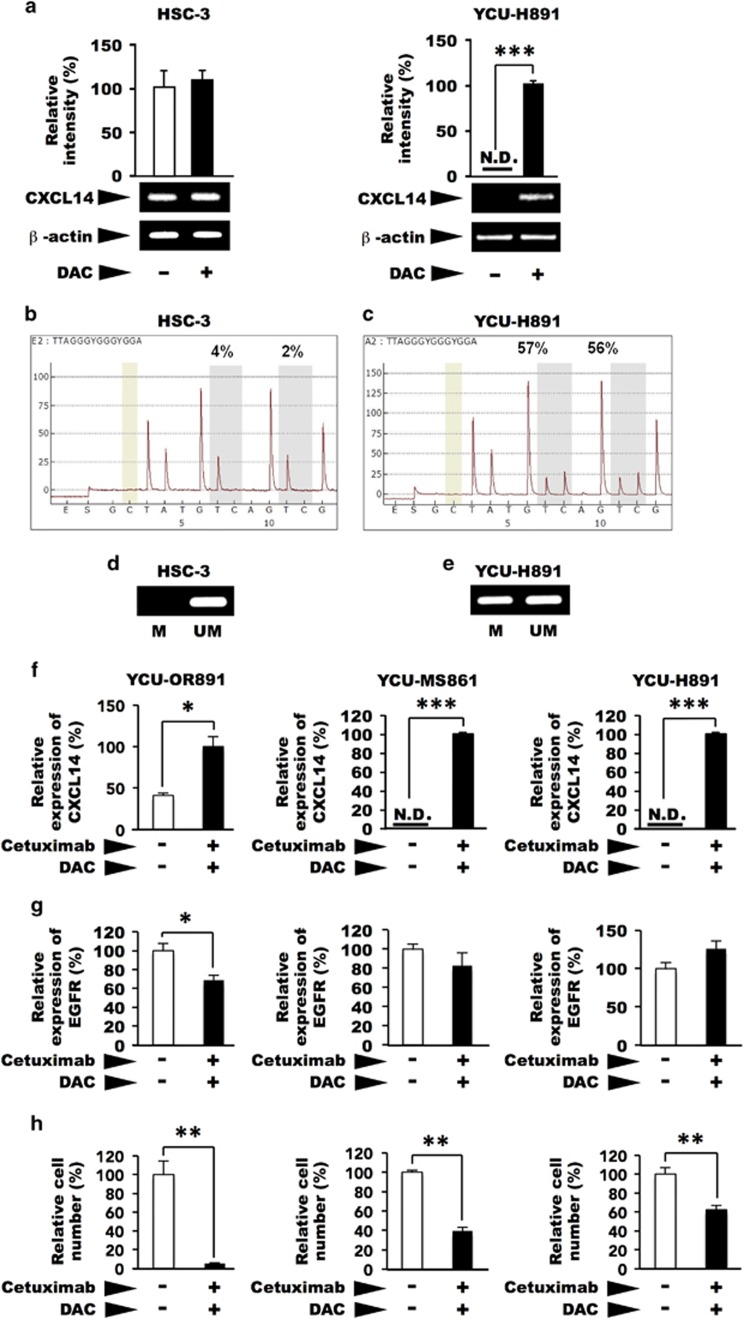
Figure 3.
Effects of 5-aza-2′-deoxycytidine(DAC) on the expression of CXCL14 mRNA and the methylation of the promoter region of CXCL14 in HSC-3 and YCU-H891 cells. (a) Beginning 1 day after the cells had been plated (1.0 × 105/60-mm dish, Corning) the medium was replaced every day with fresh medium containing DAC (Sigma-Aldrich, final concentration, 5 μM) or control. Total RNA was extracted after 3 days of culture, and the expression levels of CXCL14 were determined by reverse transcription–PCR and quantitative PCR (qPCR) or densitometry after gel electrophoresis of the complementary DNA (cDNA), as described in the Figure 1d legend. Similar levels of CXCL14 mRNA were observed in the treated and untreated control HSC-3 cells (left panel). However, CXCL14 expression was detected only in the YCU-H891 cells only in the presence of DAC (right panel). (b, c) For the methylation analysis, we seeded 1 × 105 HSC-3 and YCU-H891 cells per 60-mm dish and replaced the medium every day with fresh medium containing DAC (final concentration, 5 μM) starting on the following day. Three days after initiation of DAC treatment, we extracted RNA and verified the expression of CXCL14 and β-actin. Using a QiaAmp DNeasy kit (Qiagen), we extracted the DNA, and the EZ DNA Methylation-Gold kit (Zymo Research, Irvine, CA, USA) was used to perform bisulphite substitution. Using the PSQ assay design program (Qiagen), we designed the following PCR primers for pyrosequencing: 5′-GYG GGT TGG GAA GGT TTT-3′ (forward primer), 5′-TCR ATA AAT ACC CAA AAC TAT CT-3′ (5′-biotinylated reverse primer; PCR product size, 206 bp) and 5′-ACG AG(C/T) GGA TTT AAA AGA GG-3′ (sequencing primer). The pyrosequencing analysis was performed with a PyroMark ID system (Qiagen) and a Pyro Gold Reagent kit RRK (Qiagen). The results for the HSC-3 cells (b), and YCU-H891 cells (c) are presented. For methylation-specific PCR, we used the MethPrimer program to design PCR primers. The cycling conditions for the methylation and non-methylation PCR reactions were as follows: denaturation at 95 °C for 5 min, 45 cycles of denaturation at 95 °C for 20 s, annealing at 60 °C for 30 s and elongation at 72 °C for 30 s, followed by additional elongation at 72 °C for 5 min. (d, e) We verified the PCR products by performing agarose gel electrophoresis (2% gel). The PCR products were visualized with ethidium bromide staining after gel electrophoresis (HSC-3 cells (d) and YCU-H891 cells (e)). The values in a are expressed as the means±s.d. (n=3). Methylated (M) and unmethylated (UM) primers were used. (f, g) YCU-MS861 cells and YCU-H891 cells were inoculated into 60-mm culture dishes and cultured as described in the description in a, except that the cells were treated with 1 μg of cetuximab per ml for the last 24 h. RNA was purified by TRIzol, and qPCR was performed as described in the legend for Figure 1d. The relative rates of cDNAs for human CXCL14 (f), and human EGFR (g) are presented. For human EGFR, 5′-TCCCCGTAATTATGTGGTGAC-3′ (forward) and 5′-GCCCTTCGCACTTCTTACAC-3′ (reverse) were employed to yield a 110-bp product. (h) For the determination of the growth properties of the cells, the cells were plated in 12-well plates (Sumitomo Bakelite, Tokyo, Japan, 5 × 104 per well) and cultured as described above, except for the YCU-OR891 cells, which were inoculated at 1 × 104 per well and cultured for 6 days. Cell numbers from three wells were counted using a Coulter Z1 Counter (Coulter Electronics Ltd, UK) and relative cell numbers are presented. The values are expressed as the means±s.d. (n=3). *P<0.05, **P<0.001 and ***P<0.001 (Student's t-test). Experiments were performed in triplicate, and the values were normalized to β-actin. The figures represent one of two cell-culture experiments.