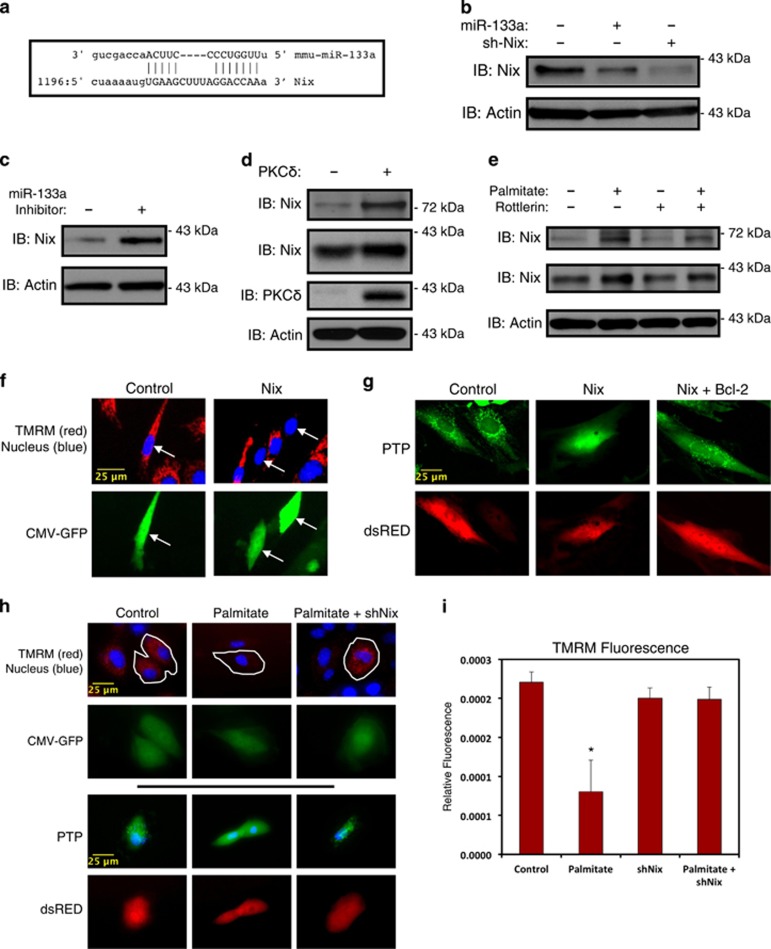
Figure 5.
miR-133a regulates mitochondrial membrane potential through Nix. (a) Sequence alignment of mouse miR-133a and the 3' UTR of Nix. (b) H9c2 myoblasts were transfected with miR-133a or a shRNA targeting Nix (sh-Nix). Following protein extraction, samples were immunoblotted as indicated. (c) H9c2 cells were transfected with a miR-133a inhibitor (50 μM) or a scrambled control oligonucleotide. Extracts were immunoblotted, as indicated. (d) C2C12 cells were transfected with the catalytic isoform of PKCδ. Extracts were immunoblotted, as indicated. (e) Five-day differentiated C2C12 cells were treated with 200 μM palmitate conjugated to 2% albumin, or 5 μM rottlerin overnight, as indicated. Protein extracts were immunoblotted, as indicated. (f) C2C12 myoblasts were transfected with Nix, or an empty vector control. CMV-GFP was included to identify transfected cells. Cells were stained with TMRM and Hoechst and imaged by standard fluorescence microscopy. Arrows indicate GFP-positive cells. (g) H9c2 cells were transfected with Nix and Bcl-2, as indicated. CMV-dsRed was used to identify transfected cells. Cells were stained with calcein-AM with cobalt chloride (5 μM) to assess PTP opening. (h) H9c2 cells were transfected with shNix or a scrambled control shRNA. Following recovery, cells were treated with 200 μM palmitate conjugated to 2% albumin overnight, and stained with TMRM and Hoechst to evaluate mitochondrial membrane potential (above) or with calcein-AM with cobalt chloride (5 μM) to assess PTP opening (below). (i) Quantification of myotube fluorescence in (h). Data are represented as mean±S.E.M. *P<0.05 compared with control