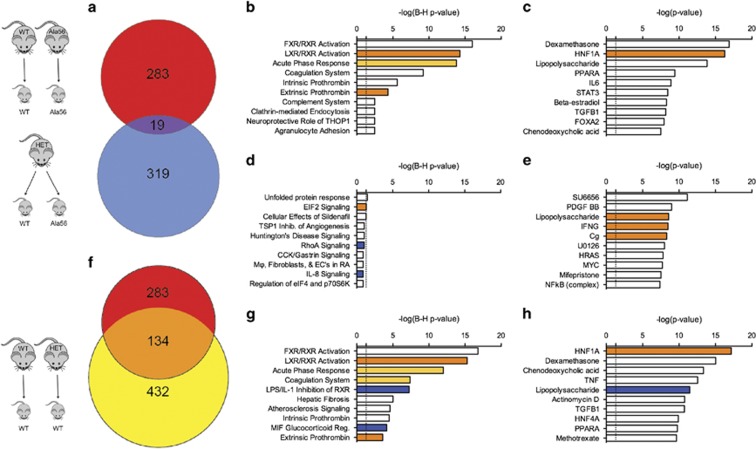
Figure 5.
Examination of maternal and embryonic SERT Ala56 genotype effects on E14.5 placental gene expression. In the homozygous and heterozygous dam comparisons, 283 and 319 differentially expressed genes (DEGs) were identified in Ala56 placentas relative to wild-type, respectively. Only 19 DEGs (a) overlapped between the two group comparisons. Conversely, of the 432 DEGs identified when comparing the wild-type placentas from heterozygous and the wild-type dams, 134 DEGs overlapped with the homozygous dam group comparison (f). Ingenuity Pathway Analysis (IPA) identified DEG-enriched canonical pathways and predicted upstream regulators, with the top 10 shown for the homozygous dam (b and c), heterozygous dam (d and e), and wild-type (g and h) data sets. Light and dark orange bars are moderately and strongly predicted upregulated pathways/networks, respectively. Blue bars are strongly predicted downregulated pathways/networks. White bars indicate that the direction of activation could not be predicted based on the direction of regulation of DEGs within the pathways/networks. Vertical dashed line indicates threshold required to meet statistical significance (p<0.05) for pathway/network enrichment.