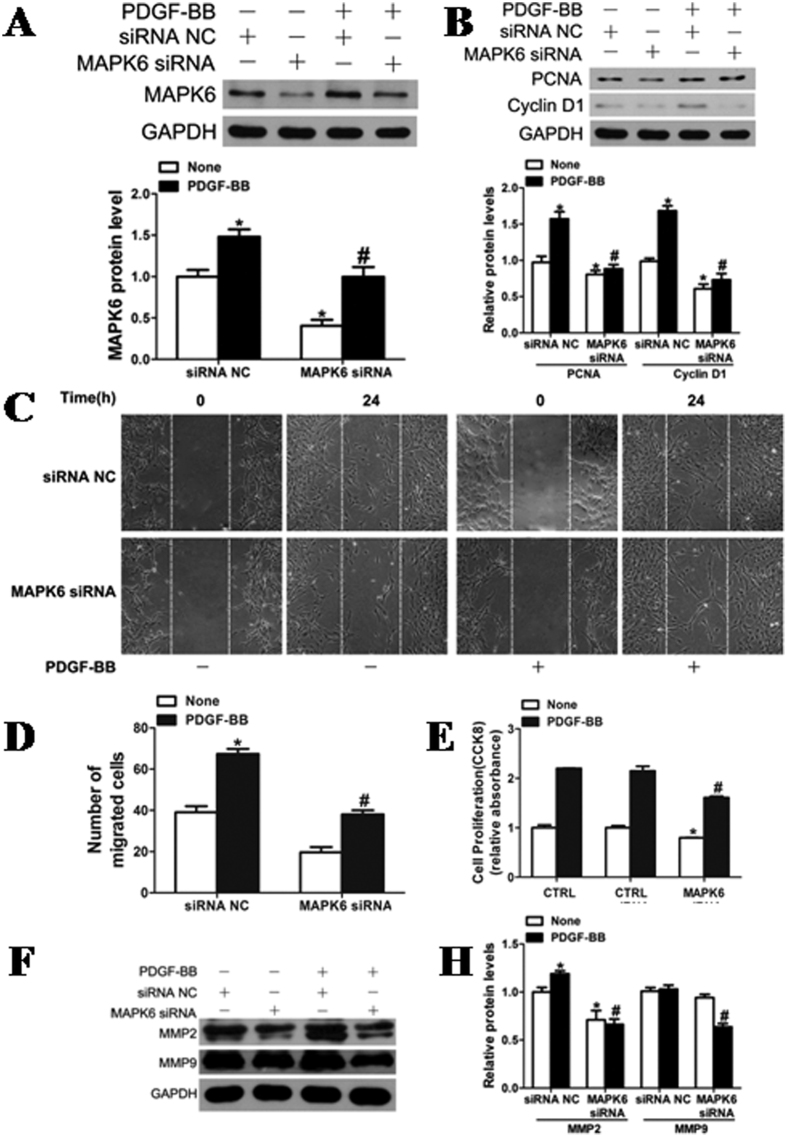
Figure 5. Role of MAPK6 in VSMC proliferation and migration.
(A) MAPK6 expression was reduced by silencing MAPK6 with specific small interfering RNA (MAPK6 siRNA; 100 nM). *P < 0.05 represents statistical significance compared with the control siRNA without PDGF-BB, #P < 0.05 represents statistical significance compared with the control siRNA with PDGF-BB. (B) The expression of PCNA and Cyclin D1 was reduced after silencing MAPK6. *P < 0.05 represents statistical significance compared with control siRNA without PDGF-BB, #P < 0.05 represents statistical significance compared with control siRNA with PDGF-BB. The original images for (A and B) can be seen in Supplementary Fig. 2. (C) VSMCs transfected with MAPK6 siRNA (100 nM) were assessed for cell migration after stimulation with PDGF-BB (20 ng/ml) for 24 h by scratch-wound assays. (D) Quantitation of migrated cells. The data are shown as the mean ± SD of the number of migrated cells from 3 independent experiments. *P < 0.05 represents statistical significance compared with the control siRNA without PDGF-BB, #P < 0.05 represents statistical significance compared with the control siRNA with PDGF-BB. (E) MAPK6 deficiency attenuated PDGF-BB (20 ng/ml)-induced proliferation of VSMCs as determined by CCK8 assays (n = 4). (F) Rat jugular vein VSMCs transfected with MAPK6 siRNA (100 nM) or siRNA NC (100 nM) were stimulated with or without PDGF-BB (20 ng/ml) for 24 h followed by analysis of MMP-2 and MMP-9 protein levels with western blotting. The original images for (F) can be seen in Supplementary Fig. 6. (H) Densitometric analysis of MMP-2 and MMP-9 protein levels as determined by western blotting. *P < 0.05 represents statistical significance compared with the control siRNA without PDGF-BB, #P < 0.05 represents statistical significance compared with the control siRNA with PDGF-BB.