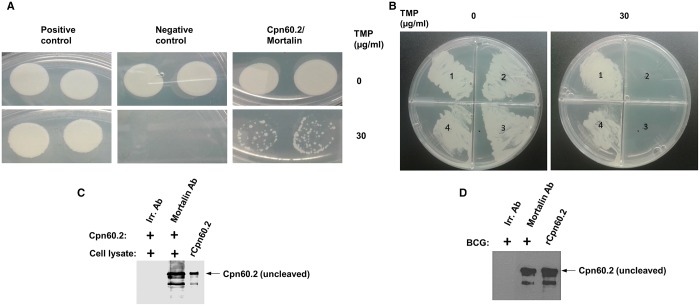
Fig. 4.
Cpn60.2 interacts with mitochondrial mortalin. (A) M. smegmatis expressing Cpn60.2-mDHFR F1,2 and mortalin-mDHFR F3 fusion proteins were plated on solid media containing trimethoprim (TMP). M. smegmatis expressing CFP-10 fused to F1,2 and ESAT-6 fused to F3 correspond to positive control. Negative control is M. smegmatis transformed with Cpn60.2-F1,2 and empty F3 constructs. The appearance of TMP-resistant bacterial colonies reflects the reconstitution of mDHFR as a result of protein-protein interaction. Experiments are shown in duplicates. (B) M. smegmatis co-transformed with Cpn60.2 (F1 and F2)/EFTM-F3 constructs and Cpn60.2 (F1 and F2)/Mortalin-F3 constructs were allowed to grow in the presence of trimethoprim as in A. M. smegmatis co-expressing Cpn60.2 (F1 and F2)/EFTM-F3 construct failed to grow in the presence of trimethoprim indicating the failure of interaction of Cpn60.2 with EFTM. (1) CFP 10[F1,2]/ESAT 6[F3], (2) Cpn60.2[F1,2]/[F3], (3) Cpn60.2[F1,2]/EFTM[F3], and (4) Cpn60.2[F1,2]/Mortalin[F3]. (C) Equal amounts of macrophage lysates were incubated with rCpn60.2 protein and subjected to pull down assays with protein A/G magnetic beads conjugated with mortalin or irrelevant antibodies. Pulled down material was then subjected to western blotting with Cpn60.2 antibody as in Fig. 1B. (D) Protein lysates from BCG-infected macrophages were subjected to immunoprecipitation with mortalin or irrelevant antibodies then analyzed along with rCpn60.2 by western blotting with Cpn60.2 antibody as in Fig. 1B. Data are representative of three independent experiments.