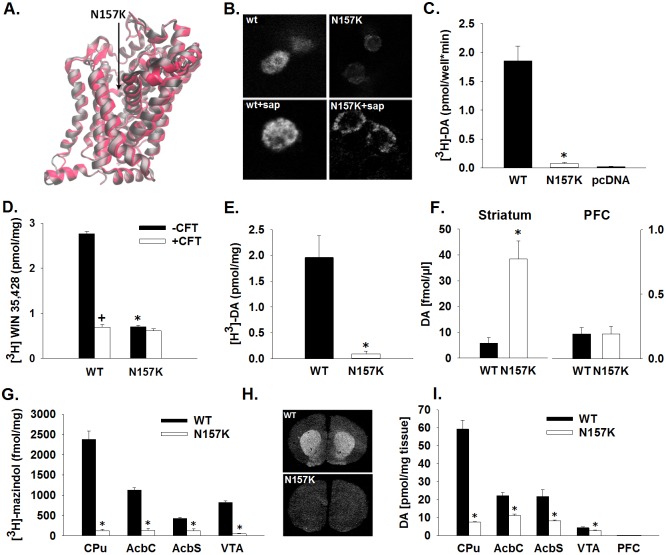
Fig. 1.
Molecular characterization of a novel mutant rat model with a subcortical hyperdopaminergic state. (A) Secondary structures of the wild-type (gray) and N157K-mutant (red) dopamine transporter (DAT) following energy minimization (1 ns) within an environment resembling the intracellular space. (B-D) Subconfluent HEK293 cells transiently transfected with rat DAT-wt (WT) or rat DAT-N157K (N157K). (B) Immunofluorescence and confocal analysis of DAT protein using an antibody targeted against an extracellular epitope of DAT in the absence (top row) and presence (bottom row) of detergent saponine (sap). A weak cell surface labeling is seen for DAT-N157K in the absence of detergent (top right) and a globular distribution of protein is seen for both DAT-wt and DAT-N157K (bottom row). Shown are example z-projections of single cells. (C) [3H]DA uptake in HEK293 cells expressing DAT. Specific [3H]DA uptake is defined as the difference between monoamine transporter mediated uptake minus control uptake in HEK293 cells that have been transiently transfected with empty vector pcDNA3.1(pcDNA). (D) Total (-CFT) and nonspecific (+CFT) binding of [3H]WIN35,428 to DAT. Nonspecific binding was determined in the presence of 50 µM β-CFT (WIN35,428). (E) Total [3H]DA uptake in synaptosomes obtained from the dorsal striatum (caudate putamen, CPu) from wild-type (WT, n=3) and DAT mutant (N157K, n=3) rats. (F) Basal extracellular DA levels in the CPu and medial prefrontal cortex (PFC) of WT (n=12) and N157K (n=9) rats. (G) Quantitative analysis of DAT ([3H]-mazindol) binding levels in the CPu, nucleus accumbens core (AcbC), nucleus accumbens shell (AcbS) and ventral tegmental area (VTA) of WT (n=10) and N157K (n=9) rats. (H) Representative dark-field images showing [3H]-mazindol binding on a coronal brain section from WT and N157K rats at the striatal level. (I) Tissue concentration of dopamine in homogenates of CPu, AcbC, AcbS, VTA and PFC of WT (n=5) and N157K (n=5) rats. All data are expressed as means±s.e.m. *P<0.05, significant difference from DAT-wt cells/WT rats; +P<0.05, significant difference from the total binding.