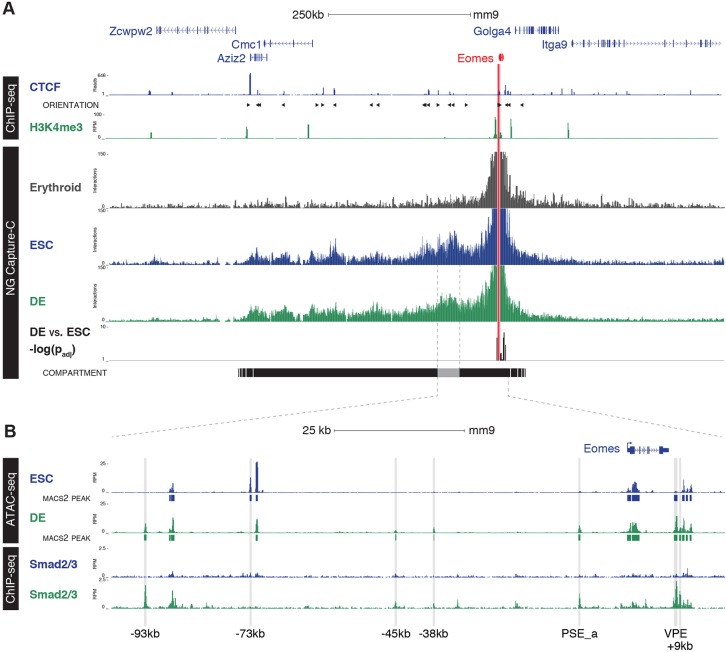
Fig. 5.
Eomes is regulated by Smad2/3 binding in a preformed compartment. (A) NG Capture-C interaction profiles of the Eomes promoter (chr9:117,683,476-118,771,067) from erythrocytes (grey), ESC (blue) and DE (green). Tracks show mean interactions of normalised biological replicates (n=3) and DESeq2 significant differences between DE and ESC [−log(Padj); P≤0.05]. The Eomes compartment, as determined by boundaries of strong promoter interactions with CTCF orientation (arrowheads), is based upon binding in ESCs (Handoko et al., 2011). Histone modifications for H3K4me3 (DE, n=3) show promoter regions. (B) Enlargement of the region of the Eomes compartment showing highest association with the promoter, from chr9: 118,252,500-118,405,500. Open chromatin was generated using ATAC-seq in ESC and DE (n=3), with the addition of MACS2 called peaks annotated beneath each ATAC-seq track and Smad2/3 ChIP-seq in ESCs (blue) and DE (green) (Yoon et al., 2015). Regions of chromatin accessibility unique to ESCs (−73 kb) and those associated with Smad2/3 occupancy in DE (−93 kb, −45 kb, −38 kb, PSE_a, VPE and +9 kb) are indicated.