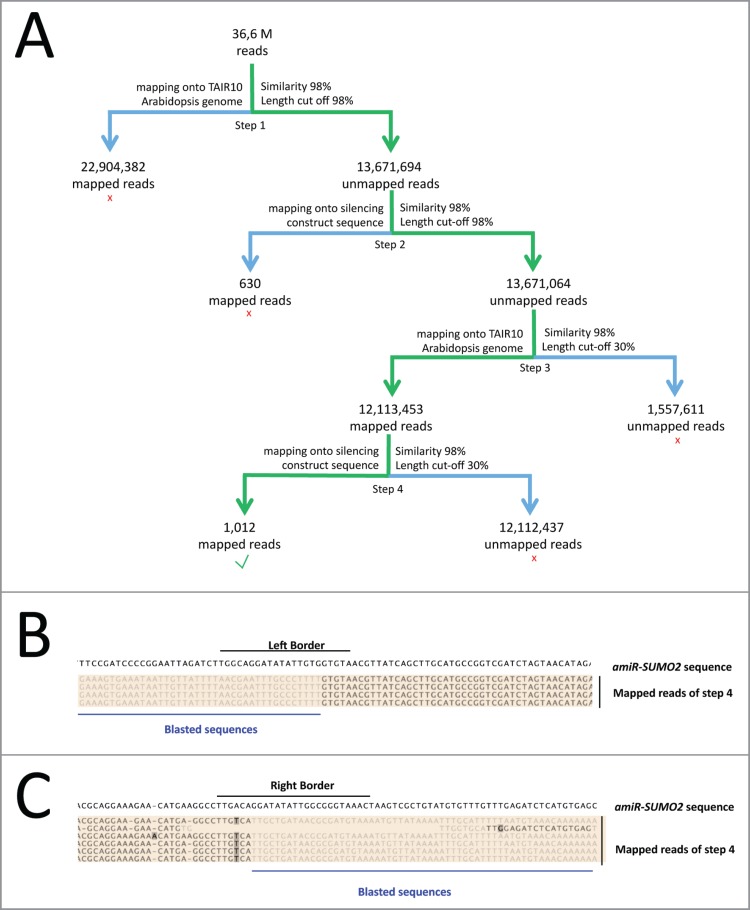
Figure 1.
Identification of the 2 amiR-SUMO2 insertion sites using NGS sequencing. (A) Pipeline used to identify the T-DNA insertion sites. (Step 1) Reads were mapped onto the Arabidopsis genome assembly (TAIR10), using a sequence similarity cut-off of > 98% and read length cut-off of > 98% sequence overlap. (Step 2) To remove the reads that fully matched to the amiR-SUMO2 construct, the unmapped reads were then mapped to the amiR-SUMO2 construct using similar parameters. (Step 3) We then selected in the remaining set of unmapped reads, the reads that partially mapped to the Arabidopsis genome, using >98% similarity and a length cut-off of > 30%. (Step 4) The retained reads (from Step 3) were then mapped to the amiR-SUMO2 construct, with >98% similarity and a length cut-off of > 30% to obtain the reads that map across single integration site boundaries with at least 30 bp. We found 1,012 reads, which mapped to 2 different genomic sites. The insertions were identified by blast searches with these latter reads using the part of the reads that did not map onto the amiR-SUMO2 construct. (B) and (C) Visualization of the mapped reads of Step 4 from panel A (shown on color background) on the SUMO2 silencing construct sequence at the Left Border (B) and Right Border (C). Within the mapped reads of step 4, black sequences indicates the regions of the reads that map onto the amiR-SUMO2 construct, while gray sequences indicate the regions of the reads from Step 4 that do not map on the amiR-SUMO2 construct.