Figure 3.
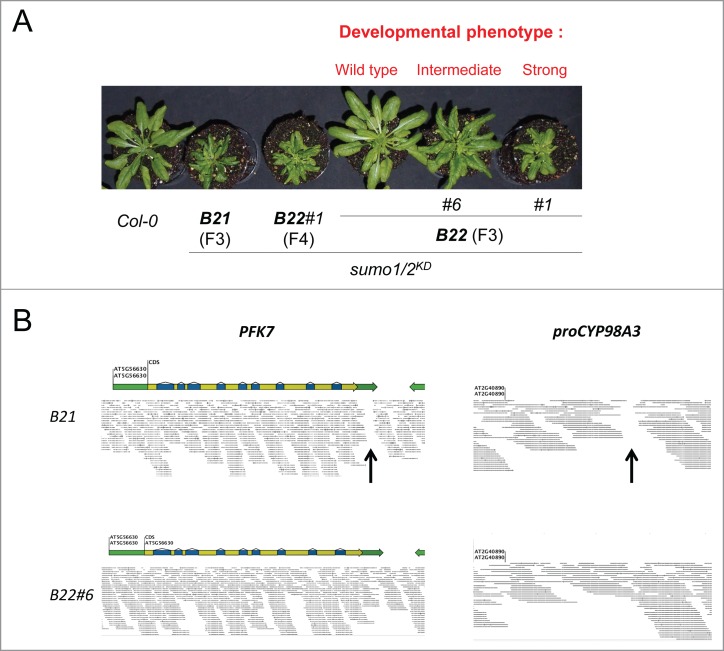
Both the amiR-SUMO2 integration events (i.e., at PFK7 and proCYP98A3) need to be present in a homozygous state to obtain a strong sumo1/2KD phenotype. (A) Seedlings from the self-pollination offspring of sumo1/2KD lines B21 (F3 generation); B22 (F3 generation); and B22#1 (F4 generations) were grown for 4 weeks in short day conditions (11 hours in day light / 13 hours in dark). The lines B21 and B22 represent 2 sister plants obtained from the same cross between sumo1–1 and SUMO2KD line B; B22#1 and B22#6 exemplify selfings from the plant B22. Whereas the strong developmental phenotype did not segregate for the progeny of B21, the progeny of B22 displayed 3 different phenotypes: normal, intermediate, and strong (i.e., ‘B21-like’). A set of B22#6 intermediate plants was pooled and their combined gDNA was sequenced using next-generation sequencing. (B) Visualization of the reads obtained from re-sequencing of the pool of the B22#6 at the PFK7 and proCYP98A3 promoter (proCYP98A3) genomic regions reveals no gap in the read coverage for both insertion sites (bottom row), while a gap in the read coverage is visible for both genomic regions in the case of the B21 plant (top row, black arrows).