Abstract
Transmembrane proteins are mostly nanochannels playing a highly important role in metabolism. Understanding their structures and functions is vital for revealing life processes. It is of fundamental interest to develop chemical devices to mimic biological channels. Structural DNA nanotechnology has been proven to be a promising method for the preparation of fine DNA nanochannels as a result of the excellent properties of DNA molecules. This review presents the development history and current situation of three different types of DNA nanochannel: tile-based nanotube, DNA origami nanochannel, and DNA bundle nanochannel.
Keywords: DNA nanotechnology, nanostructures, tile-based nanotube, origami nanochannel, bundle nanochannel
Introduction
DNA has been proven to be an excellent structural molecule besides being used for genetic information storage and transfer because the specificity of the base pairing allows target structures to be encoded in the base sequences of component oligonucleotides. After more than 30 years of development since Nadrian Seeman proposed that DNA could be used to construct junction structures in 1982 1, molecular self-assembly by DNA has enabled the construction of arbitrary objects with nanometer- to micrometer-scale dimensions (for example, crystals, patterns, bricks, boxes, and curved shapes) 2– 4. These nanostructures either perform functions independently or simulate other biomolecules; among them, the DNA nanochannel is one of the finest examples that could mimic the biological nanochannels in both structure and function. Biological nanochannels are widely distributed in nature; they play important roles in various biological processes, regulating the transport of materials and signals through biomembranes 5, 6. An accurate understanding of the structures and functions of the nanochannels would help us unveil the life processes and has important guiding significance for drug design, disease diagnosis, and manufacturing of bio-inspired devices. In this review, we will briefly introduce the development history and current situation of DNA nanochannels in three classifications: tile-based nanotube, DNA origami nanochannel, and DNA bundle nanochannel.
Tile-based nanotube
The approach of DNA tile-based structures was to use DNA to construct relatively simple tiles such as the double crossover (DX) 7 or triple crossover 8. Then, these preformed tiles could assemble into periodic lattices through DNA hybridization. The DNA tiles could be assembled into a variety of shapes; moreover, they could be used to implement algorithmic self-assembly, making them a platform for DNA computing 9. In 2004, two articles focusing on the DNA tile-based nanotube were published in the Journal of the American Chemical Society in the same issue 10, 11 ( Figure 1a). Both of these works take advantage of DX to construct the nanotubes. Owing to the periodic array of DX, the nanotubes were borderless in theory. In their experiments, both sets of investigators observed micrometer-scale nanotubes under atomic force microscope. In 2008, Yin et al. used a single-strand tile strategy to construct the DNA nanotube 12. The tile in this strategy, unlike the previous method, was only the DNA single strand. Another way to build the DNA nanotube was to use small circular DNA molecules as tiles and then introduce assisted staple strands to connect the circular DNA by forming crossover 13. Owing to the designability and biological compatibility, the tile-based nanotubes were widely used as the one-dimensional templates 14 and nanowires 15. Nevertheless, it was difficult to control their shape and size because of the periodic array of the tile, which made it hard to mimic the biological nanochannel. However, the DNA origami provided a promising way to solve this problem.
Figure 1. DNA nanochannels.
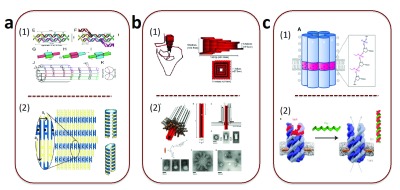
( a) Tile-based nanotube. ( b) DNA origami nanochannel. ( c) DNA bundle nanochannel.
DNA origami nanochannel
The concept of DNA origami was first proposed by Rothemund in 2006; it was assembled by folding a long single-stranded DNA into designed structures with the aid of multiple short staple strands 16. It has been widely used as a bottom-up approach for the assembly of versatile nanostructures. In 2012, Keyser et al. constructed a funnel-shaped three-dimensional DNA origami nanochannel, which could be inserted into the silicon nitride nanopores 17. The designed shape enabled the structure to fit into the solid pore size ( Figure 1b1). When the DNA origami was trapped into the solid-state nanopore, it reduced the flow of ions and therefore a characteristic drop in conductance was observed. It had been proven that hybrid DNA origami-solid-state nanopores could be used as the resistive-pulse sensor for λ-DNA. The following year, Simmel et al. published a syringe-shaped DNA origami nanochannel 18. This nanochannel could penetrate and span a lipid membrane by modifying 26 cholesterols around the channel, making the structure more closely mimic the action of biological protein nanopores ( Figure 1b2). It has also been proven that this nanochannel possessed the property of electrical conductivity by single-channel electrophysiological experiments using an integrated chip-based setup. In addition to allowing small ions to pass through, the DNA origami nanochannel could modulate molecule transport. In 2016, Krishnan et al. reported a bigger T-shaped DNA origami nanochannel incorporated into a membrane 19. This nanochannel was composed of a double-layered top plate and a 27 nm-long stem attached to the center of the plate. The stem was formed by 12 helices arranged in a square with a side length of 4.2 nm. The plate provides a large area for membrane interactions. Furthermore, Krishnan et al. had investigated an alternative strategy for inserting and anchoring DNA nanochannels into lipid membranes, which uses biotinylated lipid molecules. The specific interaction of biotinylated lipid-streptavidin and biotinylated anchor strands on DNA origami provided a different approach for interaction and methods to avoid the aggregation of DNA nanochannels. Coincidentally, Keyser et al. created a larger funnel-shaped DNA origami nanochannel the same year 20. The cross-section of the DNA nanochannel was 6 nm. Nineteen cholesterol anchors were used to facilitate insertion into a lipid membrane. Fan et al. constructed a DNA origami nanochannel with a diameter of 22 nm as the addressable bioreactor 21. They incorporated a couple of cascade enzymes into the nanochannel lumen. Compared with the free state in the solution, the nanochannel could notably enhance the cascade reaction efficiency. In 2015, Kostiainen et al. reported a modularity nanochannel based on the DNA origami strategy 22. They could controllably combine separate DNA nanochannel units equipped with a couple of cascade enzymes and demonstrate an efficient enzyme cascade reaction inside the nanochannel. The following year, Liu et al. reported a smart DNA nanochannel with a shutter at the end, which could be reversibly switched open and closed, using a DNA chain exchange reaction 23. They could regulate the transport of molecules into the nanochannel in nanoscale by controlling the state of the shutter.
DNA bundle nanochannel
Unlike DNA origami, which requires a long chain template, the DNA bundle nanochannel was very simple; it was formed by only a few oligonucleotides. This kind of nanochannel was composed of a bundle of six interconnected DNA duplexes, with an aperture of about 2 nm. The DNA bundle channel was inserted into the lipid membrane by decoration of the outer layer with hydrophobic residues 24. Howorka et al. used the hydrophobic ethyl-phosphorothioate (ethyl-PPT) DNA backbone instead of the conventional phosphate DNA backbone to construct the nanochannel 25. This way, the barrel featured a hydrophobic belt composed of 72 ethyl groups, which endowed the nanochannel with the ability of spanning the lipid membrane ( Figure 1c1). The authors proved that this nanochannel could be inserted into lipid bilayers and support a stable ionic current by single-channel current analysis. Furthermore, this DNA bundle nanochannel could interact with cellular membranes and exert a cytotoxic effect 26. Besides the capping of ethyl around the phosphate backbone, nucleoside modification was reported as a possible way to enable the insertion of the DNA nanochannels into lipid membrane. In particular, tetraphenylporphyrin (TPP) tags were introduced to the similar six-duplex bundle nanochannel, which could anchor the channel into the lipid bilayer, via the Sonogashira reaction between an acetylene-TPP derivative and a deoxyuridine nucleoside 27. Because the hydrophobicity of TPP is much stronger than that of the ethyl groups, only two TPP molecules were enough to insert the channel into the lipid membrane. In addition, the TPP possessed fluorescence emission, allowing the direct visualization of the anchored nanochannel once incubated with giant unilamellar vesicles. In 2016, Howorka et al. reported a gating nanochannel based on a similar six-bundle nanochannel 28. A lock DNA could hybridize with the channel to close it, and additional key DNA would replace the lock DNA from the channel and as a result the channel opened ( Figure 1c2). Once this gating nanochannel is inserted into the giant unilamellar vesicles with the assistance of three cholesterols, the switch of the nanochannel could regulate the release behavior of small molecules in the vesicles. Besides reporting the six-DNA bundle nanochannel, Keyser et al. reported a nanochannel composed of only four DNA bundles, which were arranged on a square lattice 29. In this arrangement, the diameter of the central channel in the middle of the four helices was only about 0.8 nm. Similarly, two cholesterols here were used to assist the channel to be inserted into the lipid membrane.
Conclusions and prospect
In this review, we have outlined the development history and current situation of the nanochannel based on the structural DNA nanotechnology. So far, three different types of DNA nanochannel have been reported: the tile-based nanotube, the DNA origami nanochannel, and the DNA bundle nanochannel. It has been proven that the DNA nanochannel was promising in the area of bio-mimic channel and molecule transport regulation of nanoscale. Furthermore, the DNA nanochannel could tune membrane fluidity and trigger a cytotoxic effect. However, this is an emerging field that in many respects is still in its infancy: one of the major challenges with the DNA nanochannels is to improve their stability for the purpose of avoiding the ionic leakage caused by fluctuations of the nanochannel in the membrane. In addition, at present, there is only one kind of responsiveness for the DNA nanochannel, and how to introduce more responsiveness into the nanochannel to better mimic the native protein channel is still a big challenge. We believe that more and more intelligent DNA nanochannels, inspired by the DNA-modified solid-state nanochannels 30, 31, which possessed different kinds of responsiveness, will be developed in the future.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Jørgen Kjems, Aarhus University, Aarhus, Denmark; Interdisciplinary Nanoscience Center (iNANO), Aarhus, Denmark
Thomas LaBean, North Carolina State University, Raleigh, NC, USA
Funding Statement
Funding was received from the National Basic Research Program of China (973 program) (grant number 2013CB932803) and the National Natural Science Foundation of China (grant numbers 91427302 and 21421064).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
References
- 1. Seeman NC: Nucleic acid junctions and lattices. J Theor Biol. 1982;99(2):237–47. 10.1016/0022-5193(82)90002-9 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. He Y, Tian Y, Ribbe AE, et al. : Highly connected two-dimensional crystals of DNA six-point-stars. J Am Chem Soc. 2006;128(50):15978–9. 10.1021/ja0665141 [DOI] [PubMed] [Google Scholar]
- 3. Zheng J, Birktoft JJ, Chen Y, et al. : From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal. Nature. 2009;461(7260):74–7. 10.1038/nature08274 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Liu M, Fu J, Hejesen C, et al. : A DNA tweezer-actuated enzyme nanoreactor. Nat Commun. 2013;4: 2127. 10.1038/ncomms3127 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 5. Gouaux E, Mackinnon R: Principles of selective ion transport in channels and pumps. Science. 2005;310(5753):1461–5. 10.1126/science.1113666 [DOI] [PubMed] [Google Scholar]
- 6. Yusko EC, Johnson JM, Majd S, et al. : Controlling protein translocation through nanopores with bio-inspired fluid walls. Nat Nanotechnol. 2011;6(4):253–60. 10.1038/nnano.2011.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu TJ, Seeman NC: DNA double-crossover molecules. Biochemistry. 1993;32(3):3211–20. 10.1021/bi00064a003 [DOI] [PubMed] [Google Scholar]
- 8. Labean TH, Yan H, Kopatsch J, et al. : Construction, Analysis, Ligation, and Self-Assembly of DNA Triple Crossover Complexes. J Am Chem Soc. 2000;122:1848–60. 10.1021/ja993393e [DOI] [Google Scholar]
- 9. Rothemund PW, Papadakis N, Winfree E: Algorithmic self-assembly of DNA Sierpinski triangles. PLoS Biol. 2004;2(12):e424. 10.1371/journal.pbio.0020424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mitchell JC, Harris JR, Malo J, et al. : Self-assembly of chiral DNA nanotubes. J Am Chem Soc. 2004;126(50):16342–3. 10.1021/ja043890h [DOI] [PubMed] [Google Scholar]
- 11. Rothemund PW, Ekani-Nkodo A, Papadakis N, et al. : Design and characterization of programmable DNA nanotubes. J Am Chem Soc. 2004;126(50):16344–52. 10.1021/ja044319l [DOI] [PubMed] [Google Scholar]
- 12. Yin P, Hariadi RF, Sahu S, et al. : Programming DNA tube circumferences. Science. 2008;321(5890):824–6. 10.1126/science.1157312 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Zheng H, Xiao M, Yan Q, et al. : Small circular DNA molecules act as rigid motifs to build DNA nanotubes. J Am Chem Soc. 2014;136(29):10194–7. 10.1021/ja504050r [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Sharma J, Chhabra R, Cheng A, et al. : Control of self-assembly of DNA tubules through integration of gold nanoparticles. Science. 2009;323(5910):112–6. 10.1126/science.1165831 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Liu D, Park SH, Reif JH, et al. : DNA nanotubes self-assembled from triple-crossover tiles as templates for conductive nanowires. Proc Natl Acad Sci U S A. 2004;101(3):717–22. 10.1073/pnas.0305860101 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Rothemund PW: Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440(7082):297–302. 10.1038/nature04586 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Bell NA, Engst CR, Ablay M, et al. : DNA origami nanopores. Nano Lett. 2012;12(1):512–7. 10.1021/nl204098n [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Langecker M, Arnaut V, Martin TG, et al. : Synthetic lipid membrane channels formed by designed DNA nanostructures. Science. 2012;338(6109):932–6. 10.1126/science.1225624 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Krishnan S, Ziegler D, Arnaut V, et al. : Molecular transport through large-diameter DNA nanopores. Nat Commun. 2016;7: 12787. 10.1038/ncomms12787 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Göpfrich K, Li CY, Ricci M, et al. : Large-Conductance Transmembrane Porin Made from DNA Origami. ACS Nano. 2016;10(9):8207–14. 10.1021/acsnano.6b03759 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 21. Fu Y, Zeng D, Chao J, et al. : Single-step rapid assembly of DNA origami nanostructures for addressable nanoscale bioreactors. J Am Chem Soc. 2013;135(2):696–702. 10.1021/ja3076692 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Linko V, Eerikäinen M, Kostiainen MA: A modular DNA origami-based enzyme cascade nanoreactor. Chem Commun (Camb). 2015;51(25):5351–4. 10.1039/c4cc08472a [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Wang D, Zhang Y, Wang M, et al. : A switchable DNA origami nanochannel for regulating molecular transport at the nanometer scale. Nanoscale. 2016;8(7):3944–8. 10.1039/c5nr08206d [DOI] [PubMed] [Google Scholar]
- 24. Hernández-Ainsa S, Keyser UF: DNA origami nanopores: developments, challenges and perspectives. Nanoscale. 2014;6(23):14121–32. 10.1039/c4nr04094e [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Burns JR, Stulz E, Howorka S: Self-assembled DNA nanopores that span lipid bilayers. Nano Lett. 2013;13(6):2351–6. 10.1021/nl304147f [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Burns JR, Al-Juffali N, Janes SM, et al. : Membrane-spanning DNA nanopores with cytotoxic effect. Angew Chem Int Ed Engl. 2014;53(46):12466–70. 10.1002/anie.201405719 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Burns JR, Göpfrich K, Wood JW, et al. : Lipid-bilayer-spanning DNA nanopores with a bifunctional porphyrin anchor. Angew Chem Int Ed Engl. 2013;52(46):12069–72. 10.1002/anie.201305765 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Burns JR, Seifert A, Fertig N, et al. : A biomimetic DNA-based channel for the ligand-controlled transport of charged molecular cargo across a biological membrane. Nat Nanotechnol. 2016;11(2):152–6. 10.1038/nnano.2015.279 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Göpfrich K, Zettl T, Meijering AE, et al. : DNA-Tile Structures Induce Ionic Currents through Lipid Membranes. Nano Lett. 2015;15(5):3134–8. 10.1021/acs.nanolett.5b00189 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Hou X, Guo W, Xia F, et al. : A biomimetic potassium responsive nanochannel: G-quadruplex DNA conformational switching in a synthetic nanopore. J Am Chem Soc. 2009;131(22):7800–5. 10.1021/ja901574c [DOI] [PubMed] [Google Scholar]
- 31. Hou X, Guo W, Jiang L: Biomimetic smart nanopores and nanochannels. Chem Soc Rev. 2011;40(5):2385–401. 10.1039/c0cs00053a [DOI] [PubMed] [Google Scholar]


