Abstract
The Rat Genome Database (RGD) (http://rgd.mcw.edu) aims to meet the needs of its community by providing genetic and genomic infrastructure while also annotating the strengths of rat research: biochemistry, nutrition, pharmacology and physiology. Here, we report on RGD's development towards creating a phenome database. Recent developments can be categorized into three groups. (i) Improved data collection and integration to match increased volume and biological scope of research. (ii) Knowledge representation augmented by the implementation of a new ontology and annotation system. (iii) The addition of quantitative trait loci data, from rat, mouse and human to our advanced comparative genomics tools, as well as the creation of new, and enhancement of existing, tools to enable users to efficiently browse and survey research data. The emphasis is on helping researchers find genes responsible for disease through the use of rat models. These improvements, combined with the genomic sequence of the rat, have led to a successful year at RGD with over two million page accesses that represent an over 4-fold increase in a year. Future plans call for increased annotation of biological information on the rat elucidated through its use as a model for human pathobiology. The continued development of toolsets will facilitate integration of these data into the context of rat genomic sequence, as well as allow comparisons of biological and genomic data with the human genomic sequence and of an increasing number of organisms.
INTRODUCTION
The Rat Genome Database (RGD) (http://rgd.mcw.edu) was originally created to provide the research community with a comprehensive and integrated collection of data from ongoing rat (Rattus norvegicus) genomic research, with particular emphasis on genetic markers and maps. Over time, it expanded to include radiation hybrid maps, strain information and comparative mapping tools. The initial mission was simple: to provide the genomics ‘infrastructure’ and create a starting point to develop a rat genomics community. Unlike other model organisms, the rat did not have a community interested in it, per se, but rather multiple, diverse communities interested in the biology of the rat as it related to human disease. The rat is arguably one of the dominant model organisms with more scientific manuscripts than any other model organism.
The rat entered the genomics scene in 1991 with the simultaneous publication of two genome scans looking for genes responsible for hypertension (1,2) and thus the rat genome project started. Over the next 8 years, significant genomic resources were built, RATMAP (http://www.ratmap.org) was launched in Sweden and the impetus for a RGD grew. The National Institutes of Health (NIH) funded a US-based rat genome database to house information from the tens of millions of research dollars invested and the RGD was released in 2000 (3). Since then, there has been a dramatic change in rat genomics—the informatic infrastructure for its genome now rivals that of the mouse and surpasses many model systems. RGD and its collaborators [RATMAP, NCBI (4), MGI (5), UCSC (6), EBI (7), RGSC (8), Baylor University (8), SWISS-PROT (9), BIND (10) and the PGA (http://pga.mcw.edu)] have built a powerful database that is meeting the genomic information needs of the research community and is evolving into a comprehensive platform for integrating the biology of the rat into the database. Here, we summarize our progress, data integration and concepts for evolving RGD into a phenome database as well as highlight some of the new tools.
Collins et al. (11) used the metaphor of a house to illustrate how the data from the Human Genome Project will serve as a foundation upon which a series of floors will be built to enable the application of genomics research to challenges in biology, health and society using informatics, technology and education as structural posts. With its strong and long history of research in the biochemical, neurological, nutritional, pharmacological and physiological components of biology and pathobiology, the rat, arguably the best ‘functionally’ characterized mammalian model system (12), will play a substantial role in enabling the Human Genome Project to fulfill its promise for the development of new therapies and cures for complex human diseases. The rat system brings a rich research portfolio to the blue print of the human genome via comparative genomics. Consequently, RGD will be dedicated to providing the research community with well-annotated data and the necessary tools for the integration, mining and comparative analyses required to understand the mechanisms of human disease, and to meet the needs of the expanding rat research community. Last year, accesses to RGD jumped 4-fold, most probably due to the release of the genomic sequence and the new data and tools that were made available.
DEVELOPMENTS
The RGD has grown considerably in terms of number of data objects, annotations using ontologies and tools for data integration. We initiated efforts to increase biological content in the database and to attach it to the genome, while maintaining our core strengths as a comprehensive database of rat genomic data and genomic tools. The RGD also progressed towards becoming a phenome-oriented database. For the sake of clarity, we define phenome to mean the comprehensive phenotypic characterization of a single species (13). However, we propose that this definition be expanded to include a comparative component. The concept of a human phenome database as described by Freimer and Sabatti (14) meets the goals outlined by Collins et al. (11) for building a platform to elucidate the mechanisms of complex diseases by layering genetic variation, pathway and phenotype data onto the genome, thus providing links among molecules, cells, tissues and whole organisms. Since experimentation is limited in humans, function and mechanisms elucidated from model organisms will be necessary in order to make full use of the human genome project. The rat's dominant role in physiology and drug development makes it a critical component of the human phenome efforts.
Data collection and integration
The data in RGD comes from a variety of sources ranging from curation of the literature and electronic curation to data submitted by members of the research community. As of September 2004, there were 6200 genes (26% of the predicted 24 000 genes), 2539 quantitative trait loci (QTLs) (708 rat, 1677 mouse and 152 human). For example, we have all 708 rat QTLs, 97% of mouse QTLs and 15% of human QTLs, 10 033 simple sequence length polymorphisms (SSLPs), 714 strains, 593 880 expressed sequence tags (ESTs), 7528 homologs and 11 203 references in the database.
Genomic data and toolsets
The availability of the genomic sequence has changed the users' needs with respect to RGD's data and tools. For example, the use of the radiation hybrid (RH) RHMAP server has declined from 15% of the tool accesses in 2001 to <1% in 2004. The ability to perform BLAST and BLAT (together accounting for 6% of all tool use) on genomic sequences, reduces the need to use RH mapping. Similarly, the use of the genetic maps decreased from 21% of the tool page accesses in 2003 to 5% in 2004. In contrast, we have seen a doubling (from 12 to 24%) in the requests for information about genes from 2003 to 2004. We anticipate that the need for gene annotation will only continue to increase for known and predicted genes. Indeed, 13% of the data accessed at RGD are ontology-related annotations and 1% for homolog data. In total, 38% of the data being accessed by the community relate to genomic sequence. We do, however, realize that genetic and RH maps (15) remain an essential underpinning of RGD and for rat research in general. Thus, RGD will continue to maintain these tools, but will prioritize implementation of tools focused on new data types.
In 2004, RGD released a series of new tools to facilitate the use and visualization of the enormous volume of genomic sequence and related biological data, including a rat genome browser built upon GBrowse (16). RGD's Gbrowse provides the following basic tracks: RGD markers, genes and QTLs, which have been integrated with UCSC (6) and Ensembl browser projects (17) via reciprocal links. It also provides biological data tracks based on Gene Ontology (GO) (18), Mammalian Phenotype (MP) Ontology, Disease Ontology (DO) and Pathway Ontology (PO) annotations. A second tool, the Gene Annotation tool (GAtool), enables researchers to obtain data from multiple sources, using a system that links gene IDs from KEGG (19), Swiss-Prot (9), Entrez Gene (20) and sequence identifiers from NCBI, Affymetrix (http://www.affymetrix.com) and The Institute for Genome Research (21). The tool can be queried through a web form and it returns and relates gene data with physiological and disease information through tabulation. The data points in the table are hyperlinked and allow the user to further retrieve data from the original sources. We anticipate that these tools will draw increased utilization as the users become familiar with them.
Comparative genomics
We made several modifications to our comparative genomics suite of tools, which remains a particular strength of RGD. Since its initial release in 2001, VCmap has remained a major tool, accounting for 14% of tool use. It is based on radiation hybrid maps of rat, human and mouse. The use of radiation hybrid maps, and the visualization of the results as genetic maps make it extremely easy to use. The ability to center the results on human, mouse or rat, depending upon the user's preference has drawn users from other research communities to RGD. VCMap also includes human cytogenic markers, allowing it to provide integration with a wider range of traits mapped in rat, mouse and human (22). The addition of QTLs to this tool is likely to increase its utility. It is now possible to explore QTLs between rat, mouse and human, based upon a genomic region in any one of the three species providing a comparative view of phenotypes and diseases. VCMap will continue to be a major tool for RGD for the next several years, because of its flexibility and ability to complement Genome Browsers with which it shares a lot of data. Therefore, we will turn it into a sequence-based tool over the next several years, while maintaining a ‘genetic map’ look.
Attaching biology to the genome
While the power of rat is in its physiological/biological characterization, attaching these types of data to the rat genome remains a challenge for several reasons. First, physiological studies are context dependent, e.g. is blood pressure measured by tail-cuff at age X the same as blood pressure measured by telemetry at age Z? What if the QTLs for these two traits overlap? Two, the data sets are not always easy to integrate, e.g. EKG or EEG data. Three, different strains and substrains show different phenotypes, and many investigators use outbred animals adding another degree of variation in the published data that requires human arbitration. Nonetheless, it is essential that RGD deploy a variety of strategies to accelerate the ability to capture these types of biological information.
Curation of strain information
RGD has become the ‘keeper’ of the strain lists for the rat community and currently houses 714 strains (485 inbred, 8 outbred, 193 congenic, 14 consomic and 14 transgenic). Strain information accounted for 9% of page accesses in 2004 and its annotation offers an immediate opportunity to add biological information to the database. Consequently, we have expanded our efforts to develop visualization and search tools to view the data. For example, we released a new tool, the ACP Haplotyper. This program ‘builds’ haplotypes from SSLPs for 48 strains characterized with nearly 5000 SSLP markers (23). As the number of single nucleotide polymorphisms (SNPs) that are genotyped in the different rat strains increase, we anticipate adding these data to the haplotypes. Also, our colleagues in the Program for Genomic Applications (PGA) at the Medical College of Wisconsin (MCW) have developed a series of tools for analyzing strain characteristics (http://pga.mcw.edu/pga-bin/strain_profile.cgi). RGD is evolving these into tools for the rat community. As the number of strains, and congenic, consomic, transgenic, knock-out, ENU mutant and cloned rats all increase, it is anticipated that this component of RGD will expand.
Quantitative trait loci
Another relatively easy means to assign biology to the genome is via QTLs. RGD has now curated all 708 rat QTLs. With a large number of genome scans from mouse and human research, it is useful to add these data into RGD. Therefore, we have loaded 1677 mouse QTLs from MGI and we have begun to load human QTLs. We will prioritize human QTL integration based on their relationships to phenotypes studied and mapped in rat, then based on location. For example, some regions of the rat genome have a high density of QTLs; we can survey regions of conserved synteny in humans to see if there are QTLs in this region for curation. Eventually, we will load all human QTLs and will maintain the curation in RGD as the genes are identified and the mechanisms of disease elucidated.
Ontologies
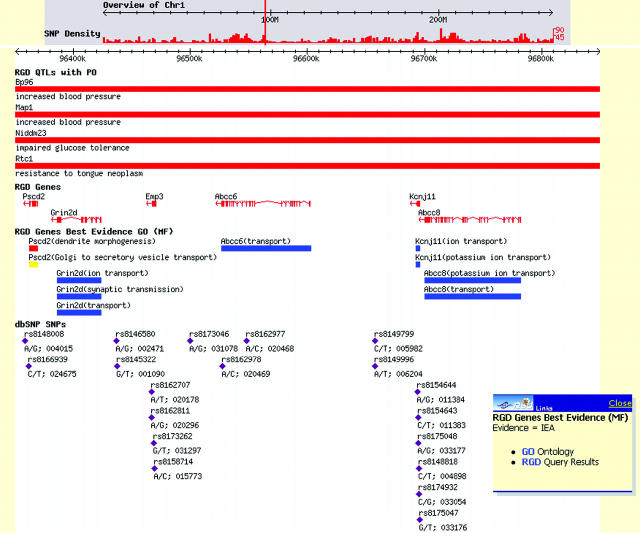
The use of ontologies provides the framework for classifying, representing and navigating across gene, phenotype and disease information, answering the community's need to link genomic data to physiology and disease (18,24). Ontologies are a powerful means of facilitating the search for information across numerous data sets. RGD implemented the GO (18), for gene data, followed by Mammalian Phenotype (MP) ontology and DO to facilitate functional annotations. The MP was initially developed at Mouse Genome Informatics (5) and is now being developed in a collaborative effort between RGD and MGI. For the DO, we adopted the ‘C’ branch of the Medical Subject Headings (MeSH) (25) hierarchy used by the National Library of Medicine (NLM). There are 65 817 gene annotations to GO in RGD: 22 093 for rat, 16 527 for human and 27 197 for mouse. For the MP, there are 2010 annotations for 388 rat genes, 836 QTLs, 384 homologs and 402 strains, while the DO has 2114 annotations (636 rat genes, 756 QTLs, 542 homologs and 180 strains). A fourth ontology for annotating pathway data has been developed at RGD in order to integrate data from existing pathway databases such as the Kyoto Encyclopedia of Genes and Genomes (19), REACTOME (26), GenMapDB (27) and the Biomolecular Interaction Database (9), as well as pathway data found in the literature. Currently, there are 373 annotations to the PO. In 2004, a new Advanced Search was released in RGD featuring Boolean methods that allow users to combine query concepts and delimiters for the types of biological data to be returned, e.g. genes, QTLs and strains. Unlike most search engines, RGD searches both ontology annotations and synonyms, and free text notes. The inclusion of ontology annotation and synonym searching provides power beyond simple pattern matching. This new tool has met with success as it now accounts for 64% of tool use, up from 38% in 2003. The large increase in the use of this tool, and the decrease in the use of other tools, suggest that the ontologies are facilitating searches and providing the users with the information they need. Direct access to RGD's ontology system is available through a browser that provides entry to all the graphs and their annotations via a search form (http://rgd.mcw.edu/ontology/ont_search.cgi). Figure 1a shows an example of an ontology report page. The ontology report provides links to detailed information (object reports) about the objects annotated with the term. The object reports, in turn, provide lists of terms to which each object is linked (Figure 1b). Through these reciprocal links, researchers can traverse related terms within and between ontologies and sets of objects, including genes, QTLs and strains. ‘Ontology tracks’ have also been implemented as browser tracks (Figure 2), some with an associated drop down menu that enable users to obtain more information on the object or the term. The tracks are intended to provide researchers with the ability to inspect the genome for biological phenomena, expressed through ontology annotations, related to specific sequence features like genes and QTLs. They can show points and regions of consequence on the genome such as exons and SNPs and relate them spatially to biological phenomena.
Figure 1.
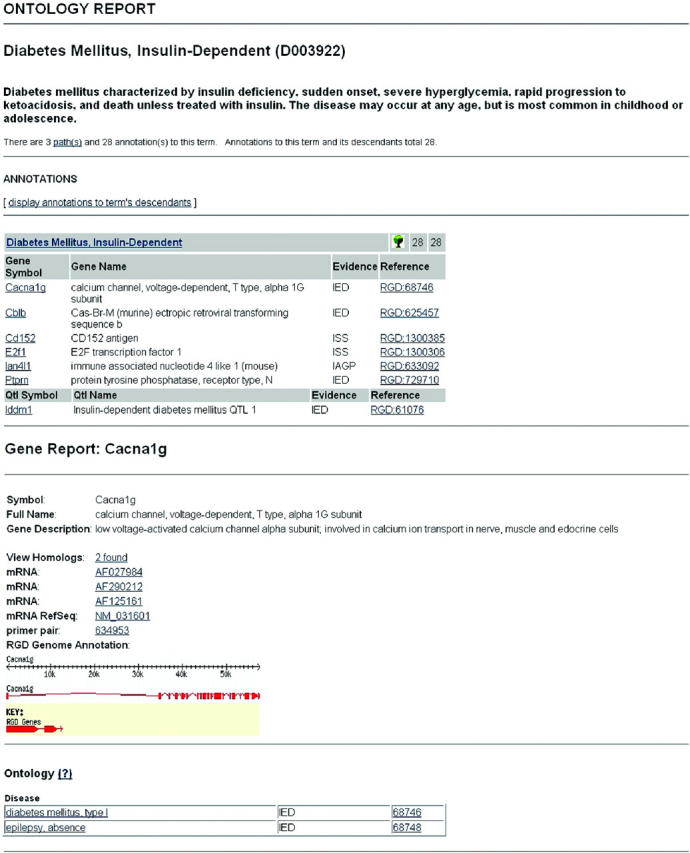
Sections of the Ontology and Gene reports. The ontology report (a) provides details on an ontology term including links to data on objects annotated to it. The gene report (b) provides links to ontology terms to which it is annotated. These reciprocal links enable users to move through various ontologies and find related terms and the objects annotated to them.
Figure 2.
A section of a map from RGD's Genome Browser showing QTL, genes and SNP tracks juxtaposed against ‘ontology tracks’. This display enables users to quickly survey biological information annotated to sequence features such as genes and QTLs and correlate the annotations with other features in the genome. The drop down menu enables the user to access multiple data reports from the ontology track. From this point, the user can obtain detailed information about the sequence feature (gene), the term to which it is annotated or the set of genes annotated to this term.
Activities within the rat community and with other databases/browsers
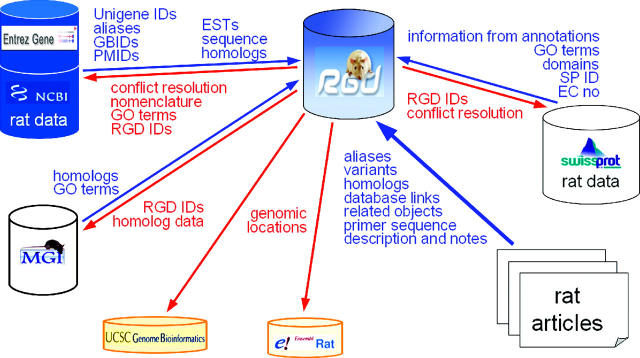
Over the last 4 years, RGD has been the communication hub for the rat community. This role remains steady and accounts for 13% of all tool use at RGD [7% for Rat Community Forum, (RCF) 1% for Newsletter and 5% for help]. Users find announcements of meetings, courses and changes to RGD. RGD has also become a major conduit for communication between the disparate rat users via the RCF. We also continue to try and expand ways for the community to participate in building the data repository through web forms for researchers to register and submit gene, QTL and strain data. RGD works with the Mouse Genome Informatics (5) and Human Genome Organization (28) to achieve uniformity and standardization in gene nomenclature and to enable the rat, human, mouse and other biomedical research communities to relate discoveries within each other's domains. Moreover, RGD has numerous links with other databases and browsers, which represent many of our collaborators. Figure 3 illustrates a few of these relationships and denotes RGD's role in ensuring that rat data are accurate and available for these groups. Finally, RGD seeks to establish important reciprocal links to other major sites to facilitate navigation between the various groups that house, generate and analyze data and information referenced or useful to RGD users.
Figure 3.
A schematic diagram illustrating the flow of data between RGD and our collaborators. More than just an exchange of information, a large amount of RGD data that has been curated and quality controlled finds its way to other major databases and genome browsers.
DISCUSSION
The RGD mission to provide researchers with current genetic and genomic data, as well as the tools necessary to relate these data and information to the physiology and biology of complex phenotypes and diseases, is on task. We expanded our efforts to meet the evolving needs of the research community. As demonstrated here, we increased the biological content of RGD, explored new ways to manage and query data and made curation more efficient. Our genomics and genetic data sets continue to expand, as do the tools that enable the users to capitalize on the data.
RGD remains sensitive to the community's needs and prioritizes data curation, tool and ontology development and collaborative activities accordingly. For example, currently, there is an expressed need to develop a system for managing the genomic sequence for the rat community. While the draft rat genome sequence is 6.8X and its assembly is very good, there are no plans to finish it. Consequently, there will be a need to update the genomic sequence assembly as further research reveals inaccuracies. Given RGD's role in the rat community, it seems natural for it to fill this critical need. We are working with the genome sequence group at Baylor University to develop a system for providing continued annotation updates of the sequence.
Future developments
RGD plans to annotate all genes in the rat over the next couple of years, and to convert the VCMap tool into a sequence-based tool. We anticipate RGD will serve as a ‘clearing house’ for changes to the genomic sequence as the community continues to use the data. We anticipate that results from most molecular genetics approaches will be actively tied to the genomic sequence. It will serve as a Rosetta stone between the human sequence and those of other model organisms. The curation teams at RGD have also identified new target information for incorporation, including mutant genes, alleles and full-length cDNA data, along with details on mutant, knock-out, congenic and transgenic strains.
Knowledge representation through ontology annotations will be made more sophisticated through data mining of the orthogonal relationships between the different ontologies. The resulting information should shed light on the relationships between the knowledge bases that they anchor and on the information within them. These developments will provide a more sophisticated infrastructure for the ontology system. Given that RGD's ontologies cover molecular, phenotypic and disease data, this is another avenue for bridging the distance between genomics and biomedicine.
Finally, we are continuously working towards making interactions with the RGD easier and more productive. Tools that provide visual cues to help navigate the data are under development. An infrastructure that allows the outputs of tools and reports to be input directly to other tools is also planned. A prototype system that sends queries to the Gene Annotation tool from the ontology system is being implemented. Furthermore, recognizing the diversity of communities of rat researchers, portal and portlet technologies, which allow users to customize the presentation of web data according to their needs, are also earmarked for implementation.
Acknowledgments
ACKNOWLEDGEMENTS
RGD is funded by grant HL64541 (H.J.) from the following institutes and centers on behalf of the NIH: the National Heart, Lung and Blood Institute, the National Eye Institute, the National Institute on Aging, the National Cancer Institute, the National Institute on Drug Abuse, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Dental and Craniofacial Research, the National Institute of Environmental Health Sciences, the National Human Genome Research Institute, the National Institute on Deafness and Other Communication Disorders, the National Institute on Alcohol and Alcoholism, the National Institute of Child Health and Human Development, the National Institute of Mental Health, the National Center for Research Resources and the National Institute of Neurological Disorders and Stroke.
REFERENCES
- 1.Hilbert P., Lindpaintner,K., Beckmann,J.S., Serikawa,T., Soubrier,F., Dubay,C., Cartwright,P., De Gouyon,B., Julier,C., Takahasi,S. et al. (1991) Chromosomal mapping of two genetic loci associated with blood-pressure regulation in hereditary hypertensive rats. Nature, 353, 521–529. [DOI] [PubMed] [Google Scholar]
- 2.Jacob H.J., Lindpaintner,K., Lincoln,S.E., Kusumi,K., Bunker,R.K., Mao,Y.P., Ganten,D., Dzau,V.J. and Lander,E.S. (1991) Genetic mapping of a gene causing hypertension in the stroke-prone spontaneously hypertensive rat. Cell, 67, 213–224. [DOI] [PubMed] [Google Scholar]
- 3.Twigger S., Lu,J., Shimoyama,M., Chen,D., Pasko,D., Long,H., Ginster,J., Chen,C.F., Nigam,R., Kwitek,A. et al. (2002) Rat Genome Database (RGD): mapping disease onto the genome. Nucleic Acids Res., 30, 125–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ostell J.M., Wheelan,S.J. and Kans,J.A. (2001) The NCBI data model. Methods Biochem. Anal., 43, 19–43. [DOI] [PubMed] [Google Scholar]
- 5.Eppig J.T., Bult,C.J., Kadin,J.A., Richardson,J.E., Blake,J.A. and the Mouse Genome Database Group (2005) The Mouse Genome Database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res., 33, D471–D475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karolchik D., Baertsch,R., Diekhans,M., Furey,T.S., Hinrichs,A., Lu,Y.T., Roskin,K.M., Schwartz,M., Sugnet,C.W. and Thomas,D.J. (2003) The UCSC Genome Browser Database. Nucleic Acids Res., 31, 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rodriguez-Tome P. (2001) EBI databases and services. Mol. Biotechnol., 18, 199–212. [DOI] [PubMed] [Google Scholar]
- 8.The Rat Genome Project Sequencing Consortium (2004) Genome sequence of the brown norway rat yields insight in mammalian evolution. Nature, 428, 493–521. [DOI] [PubMed] [Google Scholar]
- 9.Bairoch A., Apweiter,R., Wu,C.H., Barker,W.C., Boeckmann,B., Ferro,S., Gasteiger,E., Huang,H., Lopez,R., Magrane,M. et al. (2005) The universal protein Resource (UniProt). Nucleic Acids Res., 33, D154–D159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bader G.D., Betel,D. and Hogue,C.W. (2003) BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res., 31, 248–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins F.S., Green,E.D., Guttmacher,A.E. and Guyer,M.S. (2003) A vision for the future of genomics research. Nature, 422, 835–847. [DOI] [PubMed] [Google Scholar]
- 12.Hedrich H.J. (2000) History, strains and models. In Krinke,G. (ed.), The Laboratory Rat. Academic Press, London, pp. 3–8. [Google Scholar]
- 13.Mahner M. and Kary,M. (1997) What exactly are genomes, genotypes and phenotypes? and what about phenomes? J. Theor. Biol., 186, 55–63. [DOI] [PubMed] [Google Scholar]
- 14.Freimer N. and Sabatti,C. (2003) The Human phenome project. Nature Genet., 34, 15–21. [DOI] [PubMed] [Google Scholar]
- 15.Kwitek A.E., Gullings-Handley,J., Yu,J., Carlos,D.C., Orlebeke,K., Nie,J., Eckert,J., Lemke,A., Andrae,J.W., Bromberg,S., Pasko,D. et al. (2004) High density rat radiation hybrid maps containing over 24 000 SSLPs, genes and ESTs provide a link to the rat genome sequence. Genome Res., 14, 750–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stein L.D., Mungall,C., Shu,S., Caudy,M., Mangone,M., Day,A., Nickerson,E., Stajich,J.E., Harris,T.W., Arva,A. and Lewis,S. (2002) The generic genome browser: a building block for a model organism system database. Genome Res., 12, 1599–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T. et al. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ashburner M. and Lewis,S. (2002) On ontologies for biologists: the Gene Ontology—untangling the web. Novartis Found. Symp., 247, 66–80. [PubMed] [Google Scholar]
- 19.Kanehisa M. (2002) The KEGG database. Novartis Found. Symp., 247, 91–101. [PubMed] [Google Scholar]
- 20.Schuler G.D., Epstein,J.A., Ohkawa,H. and Kans,J.A. (1996) Entrez: molecular biology database and retrieval system. Methods Enzymol., 266, 141–162. [DOI] [PubMed] [Google Scholar]
- 21.Quackenbush J., Cho.J., Lee,D., Liang,F., Holt,I., Karamycheva,S., Parvizi,B., Pertea,G., Sultana,R. and White,J. (2001) The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res., 29, 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Twigger S.N., Nie,J., Ruotti,V., Yu,J., Chen,D., Li,D., Mathis,J., Narayanasamy,V., Gopinath,G.R., Pasko,D. et al. (2004) Intergrative genomics: in silico coupling of rat physiology and complex traits with mouse and human data. Genome Res., 14, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steen R.G., Kwitek-Black,A.E., Glenn,C., Gullings-Handley,J., Van Etten,W., Atkinson,O.S., Appel,D., Twigger,S., Muir,M., Mull,T. et al. (1999) A high-density integrated genetic linkage and radiation hybrid map of the laboratory rat. Genome Res., 9, AP1–8, insert. [PubMed] [Google Scholar]
- 24.Stevens R., Goble,C.A. and Bechhofer,S. (2000) Ontology-based knowledge representation for bioinformatics. Brief Bioinformatics, 1, 398–414. [DOI] [PubMed] [Google Scholar]
- 25.Nelson S.J., Johnston,D. and Humphreys,B.L. (2001) Relationships in Medical Subject Headings. In Bean,C.A. and Green,R. (eds), Relationships in the Organization of Knowledge. Kluwer Academic Publishers, NY, pp. 171–184. [Google Scholar]
- 26.Joshi-Tope G., Vastrik,I., Gopinath,G.R., Matthews,L., Schmidt,E., Gillespie,M., D'Eustachio,P., Jassal,B., Lewis,S., Wu,G. et al. (2003) The genome knowledgebase: a resource for biologists and bioinformaticists. Cold Spring Harb. Symp. Quant. Biol., 68, 237–243. [DOI] [PubMed] [Google Scholar]
- 27.Dahlquist K.D., Salomonis,N., Vranizan,K., Lawlor,S.C. and Conklin,B.R. (2002) GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genet., 31, 19–20. [DOI] [PubMed] [Google Scholar]
- 28.Bodmer W.F. (1991) HUGO: the Human Genome Organization. FASEB J., 5, 73–74. [DOI] [PubMed] [Google Scholar]