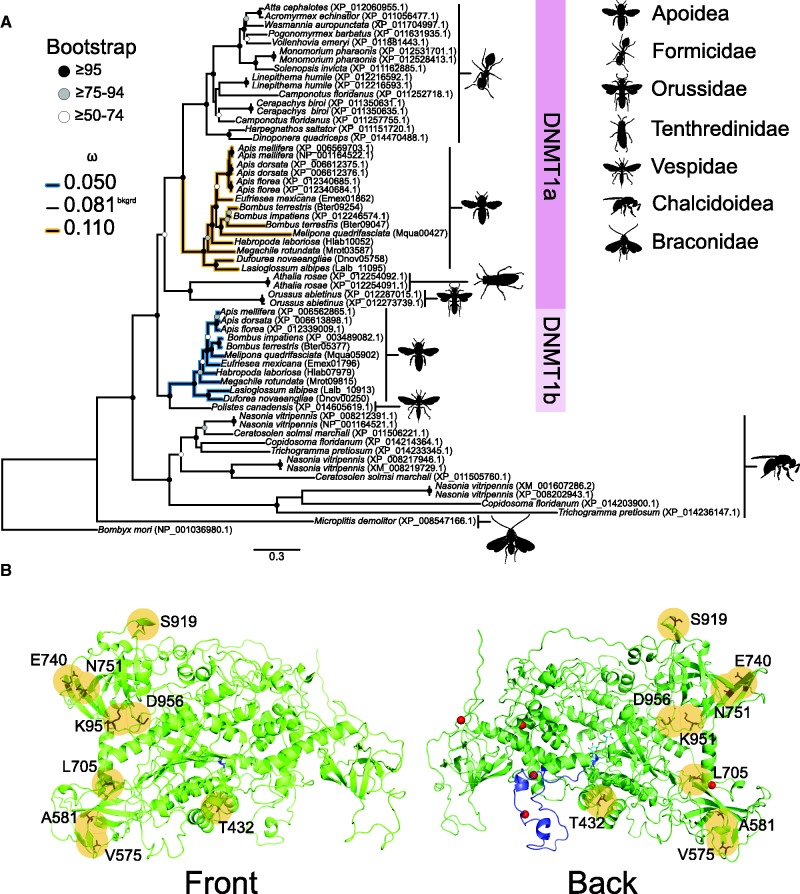
Fig. 4.
Divergent nonneutral evolution of DNMT1a and DNMT1b in Apoidea. (A) Hypothesized relationships among DNMT1 in Hymenoptera suggests a duplication event shared by the superfamilies Apoidea (bees) and family Formicidae (ants) gave rise to what is referred to as DNMT1a and DNMTb (supplementary fig. S3, Supplementary Material online). DNMT1b appears to have been lost from Formicidae (ants), whereas both DNMT1a and DNMT1b were retained in Apoidea (families Apidae, Halictidae, and Megachilidae). Divergent selection between DNMT1a and DNMT1b in Apoidea suggests the former is under relaxed purifying selection and the latter is under purifying selection. Bombyx mori (Lepidoptera) was used to root the tree, and was excluded from PAML analyses. (B) Several sites in DNMT1b were identified as under positive selection (yellow circles), with one site (T432) in the CXXC zinc finger domain, and three (V575, A581, S919) in the BAH domain. The crystal structure of Apis mellifera DNMT1b was predicted from Mus musculus DNMT1 using the program HHpred with default settings (Hildebrand et al. 2009). Details including red spheres, dark and light blue colouring specify Zn2+ ions, CXXC domain and s-adenosyl-l-homocysteine, the secondary product from the DNA methylation reaction it performs, according to Mus musculus DNMT1, respectively. Bootstrap support values are characterized as shaded circles. dN/dS (ω) values are for the most preferred branch model.