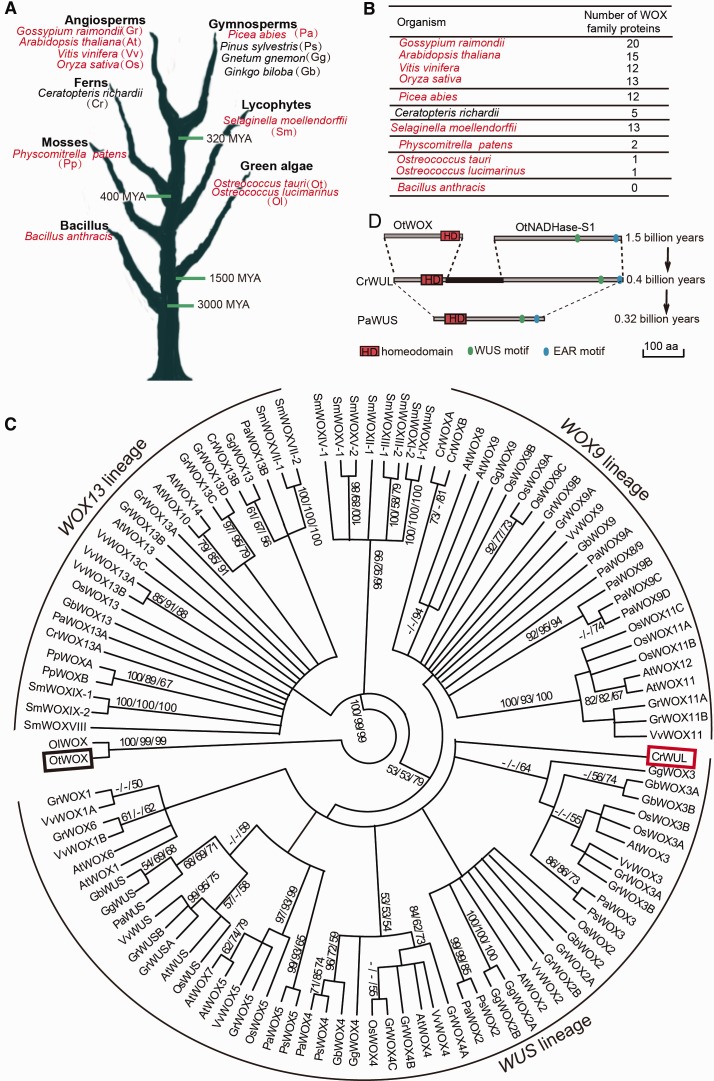
Fig. 1.
Evolution and origin of the WUS/WOX5 genes. (A) Summary of the investigated plant species, from green algae to angiosperms, according to evolutionary history. Red indicates species whose genomes have been fully sequenced. (B) Number of WOX proteins in various organisms. The distribution of the WOX members showed that the number of the WOX genes is substantially increased in the vascular plants (i.e., lycophytes, ferns, gymnosperms, and angiosperms) as compared with the nonvascular plants (i.e., mosses and green algae). (C) Phylogenetic analysis of the WOX family in the plant kingdom. The tree was divided into three lineages: the WUS lineage, the WOX9 lineage, and the WOX13 lineage. The black box highlights the single WOX identified in the green alga O. tauri. The red box highlights the single copy of a WUS-like gene, belonging to the WUS lineage, in the fern Ceratopteris richardi (CrWUL). A phylogenetic analysis was performed using the NJ/MP/ML method with full-length protein sequences. Bootstrap values are shown on the branch points. Ot, Ostreococcus lucimarinus; Ot, Ostreococcus tauri; Pp, Physcomitrella patens; Sm, Selaginella moellendorffii; Cr, Ceratopteris richardii; Gb, Ginkgo biloba; Gg, Gnetum gnemon; Ps, Pinus sylvestris; Pa, Picea abies; Os, Oryza sativa; Vv, Vitis vinifera; At, Arabidopsis thaliana; Gr, Gossypium raimondii. (D) Alignment of CrWUL with OtWOX and OtNADHase-S1 indicates that the three conserved domains common to WUS/WOX5 might have originated in fern via gene fusion. The PaWUS protein from the gymnosperm P. abies is substantially smaller than the fern CrWUL. Scale bars: 100 amino acids (aa).