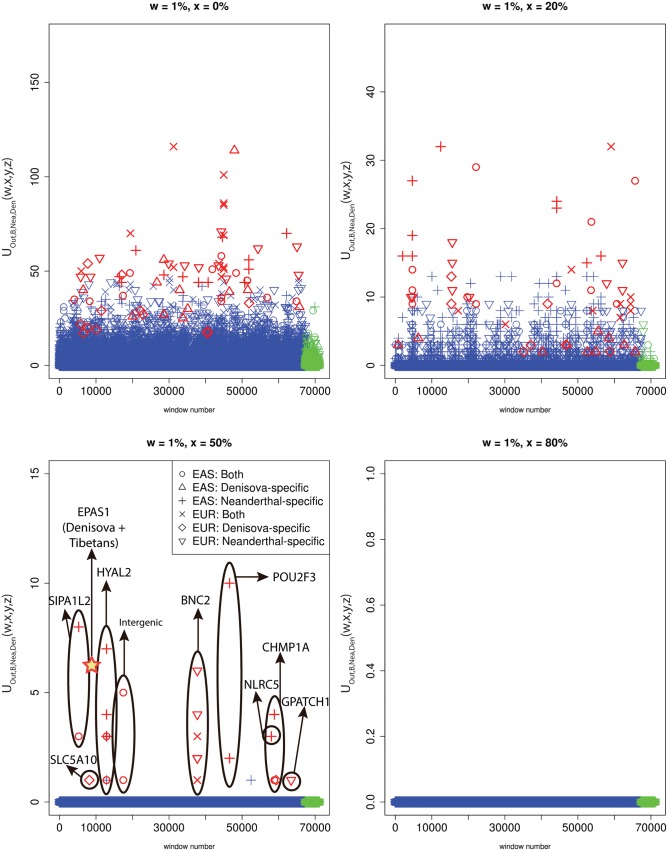
Fig. 7.
We partitioned the genome into non-overlapping windows of 40 kb. Within each window, we computed and , where Out = EAS + AFR for EUR as the target introgressed population, and Out = EUR + AFR for EAS as the target introgressed population. We searched for Neanderthal-specific alleles (), Denisovan-specific alleles () and alleles present in both archaic genomes () that were uniquely shared with either EUR or EAS at frequencies above different cutoffs (x = 0%, x = 20%, x = 50%, and x = 80%). Windows that fall within the upper tail of the distribution for each modern-archaic population pair are colored in red (P < 0.001/number of pairs tested) and those that do not are colored in blue, except for those in the X chromosome, which are in green. Ovals drawn around multiple points contain multiple windows with uniquely shared alleles that are contiguous. For comparison, the number of high frequency uniquely shared sites between Denisova and Tibetans is also shown (Huerta-Sánchez et al. 2014), although Tibetans are not included in the 1000 Genomes data and the region is 32 kb long, so this may be an underestimate.