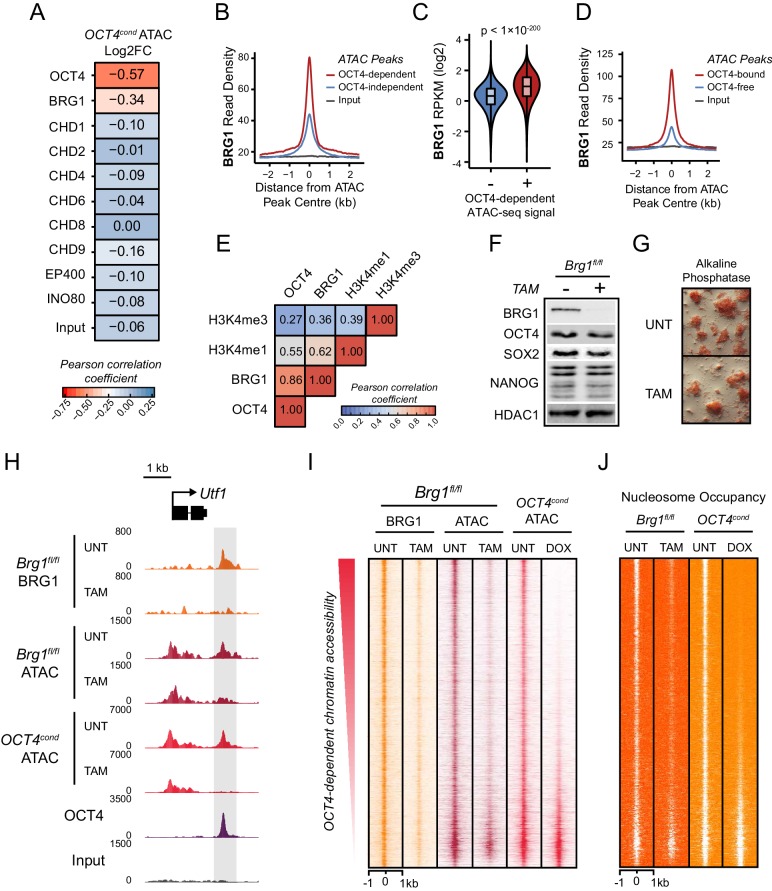
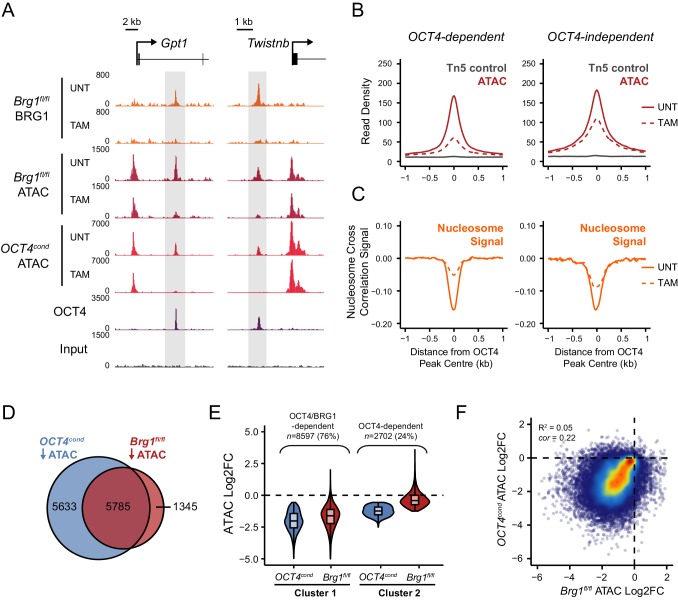
Figure 3. The chromatin remodelling enzyme BRG1 is required to create accessible chromatin at OCT4 target sites.
(A) A Pearson correlation matrix comparing log2 fold change in ATAC-seq signal in OCT4-depleted cells with wild type ESC ChIP-seq signal for nine chromatin remodellers at wild type ATAC hypersensitive peaks (n = 76,642). (B) A metaplot of BRG1 ChIP-seq signal at ATAC hypersensitive peaks with (OCT4-dependent) or without (OCT4-independent) significant reduction in ATAC-seq signal following removal of OCT4. (C) A violin plot quantifying and comparing BRG1 ChIP-seq reads per kilobase per million (RPKM) at OCT4-dependent or OCT4-independent ATAC-seq peaks depicted in (B). (D) A metaplot of BRG1 ChIP-seq signal at OCT4-bound or OCT4-free ATAC-seq peaks. (E) Genome-wide correlation of OCT4, BRG1, H3K4me1 and H3K4me3 in 2 kb windows reveals a high degree of co-localization between OCT4 and BRG1. (F) Western blot analysis for the indicated proteins in Brg1fl/fl mouse ESCs before (UNT) and after 72 hr tamoxifen (TAM) treatment. (G) Alkaline phosphatase staining of Brg1fl/fl ESCs before (UNT) and after 72 hr TAM treatment. (H) A genomic snapshot of BRG1 ChIP-seq and ATAC-seq in Brg1fl/fl ESCs before (UNT) and after 72 hr TAM treatment at the distal OCT4 target site downstream of Utf1 (highlighted in grey). The OCT4cond ATAC-seq is included for comparison and reveals a co-dependency on OCT4 and BRG1 for normal chromatin accessibility. (I) A heat map of BRG1 ChIP-seq and ATAC-seq at OCT4 target sites (n = 15920) in Brg1fl/fl ESCs before (UNT) and after 72 hr TAM treatment. Sites are ranked by loss of ATAC-seq signal following removal of OCT4, as in Figure 1F, and the OCT4cond ATAC-seq is included for comparison. (J) As in (I), changes in nucleosome occupancy before (UNT) and after (TAM) BRG1 depletion are plotted based on nucleosome signal derived from the NucleoATAC package.