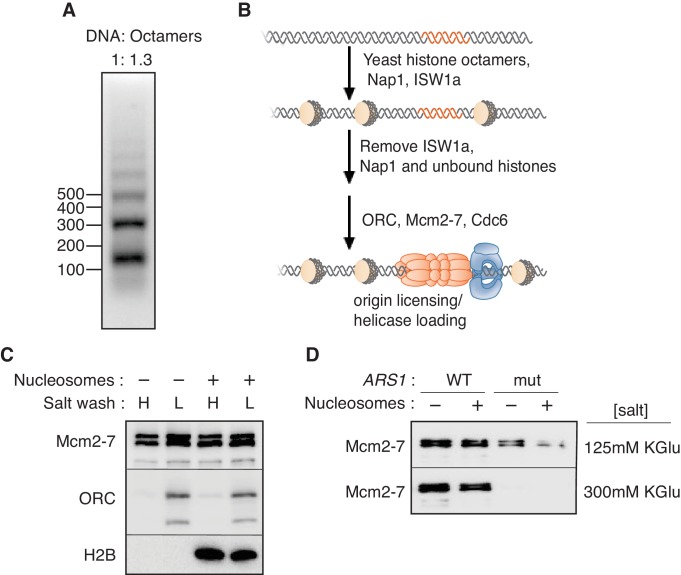
Figure 1. Mcm2-7 helicase loading onto nucleosomal DNA templates.
(A) Nucleosomes were remodeled with bead-coupled ARS1-containing linear DNA, ISW1a, yeast histone octamers and Nap1. Nucleosome assembly was assessed after partial MNase digestion. (B) Outline of the helicase-loading assay using nucleosomal DNA. (C). Comparison of helicase loading on naked DNA and on ISW1a-remodeled nucleosomal DNA. DNA templates were washed with high-salt (H) or low-salt (L) buffer after loading. Template-associated Mcm2-7, ORC and H2B was detected by immunoblot. (D) Helicase loading onto either wild-type (WT) or A-B2- (mut) (Heller et al., 2011) ARS1-containing DNA. As indicated, nucleosomal DNA was remodeled with ISW1a. Assays were performed in either 125 mM (to allow increased origin non-specific helicase loading) or 300 mM (origin specific helicase loading) potassium glutamate. After a high salt wash, DNA-associated Mcm2-7 was detected by immunoblot.