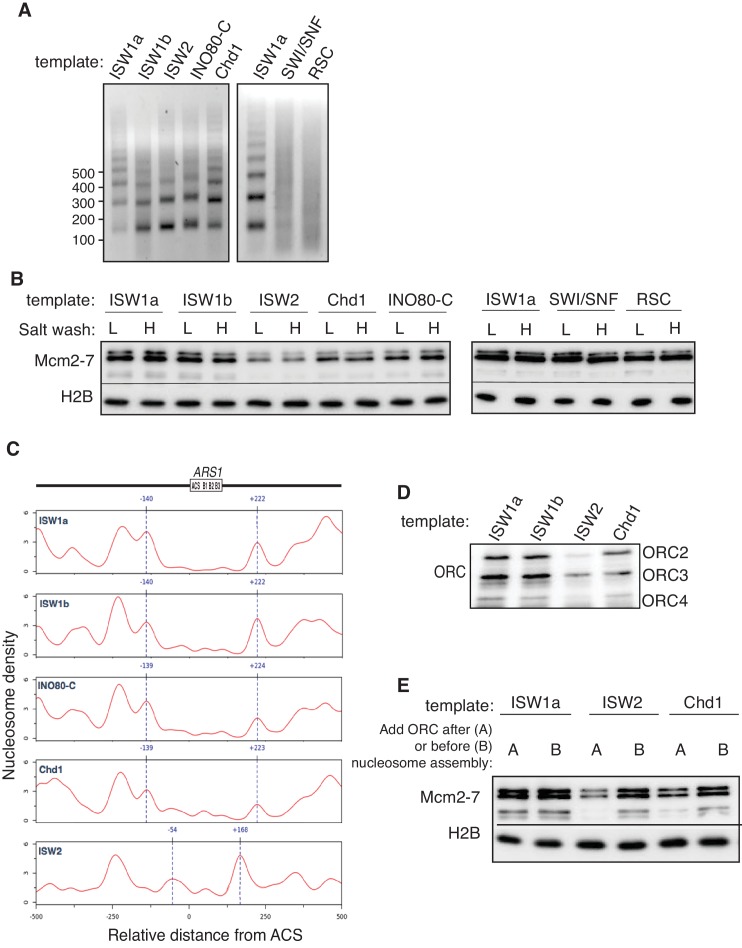
Figure 2. Comparison of helicase loading onto nucleosomal DNA templates remodeled with different CREs.
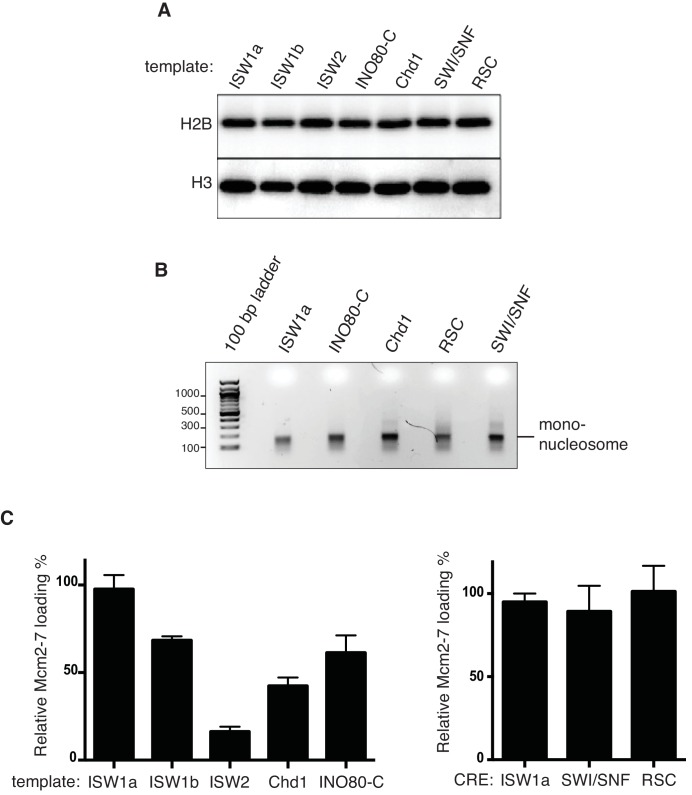
(A) Comparison of nucleosome assembly with different CREs. Nucleosomes were remodeled with the indicated CRE and assayed by partial MNase digestion. (B) Helicase loading onto nucleosomes remodeled with different CREs. After helicase loading, DNA was washed either with high-salt (H) or low-salt (L) buffer. Mcm2-7 and H2B DNA association was detected by immunoblot. (C) Comparison of origin-proximal nucleosome positioning established by different CREs. The positions of nucleosome dyads remodeled with the indicated CRE were analyzed by high-throughput MNase-Seq. Nucleosome dyad density (Y-axis) and the corresponding position of the dyad (X-axis) are plotted. Zero on the X-axis indicates the first nucleotide of the ARS1 consensus sequence (ACS). The elements of ARS1 (Marahrens and Stillman, 1992) are indicated above. (D) ORC association with nucleosomal DNA remodeled with different CREs. Template association of ORC was detected by immunoblot. (E) Addition of ORC during nucleosome assembly restores helicase loading on ISW2 and Chd1 templates. Nucleosomes were assembled onto ARS1 DNA with the indicated CRE in the presence or absence of ORC. Helicase loading was performed and analyzed as described in (B).