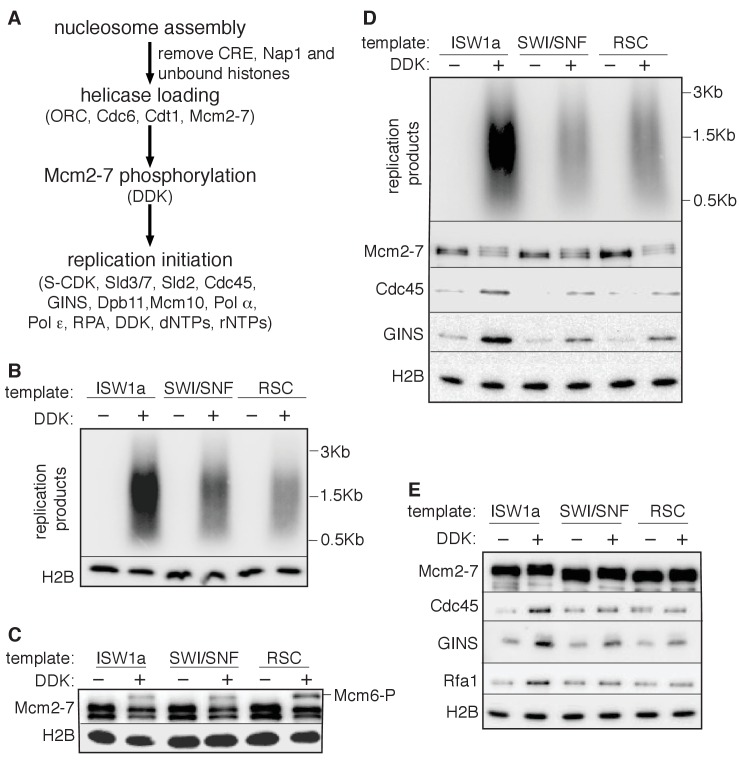
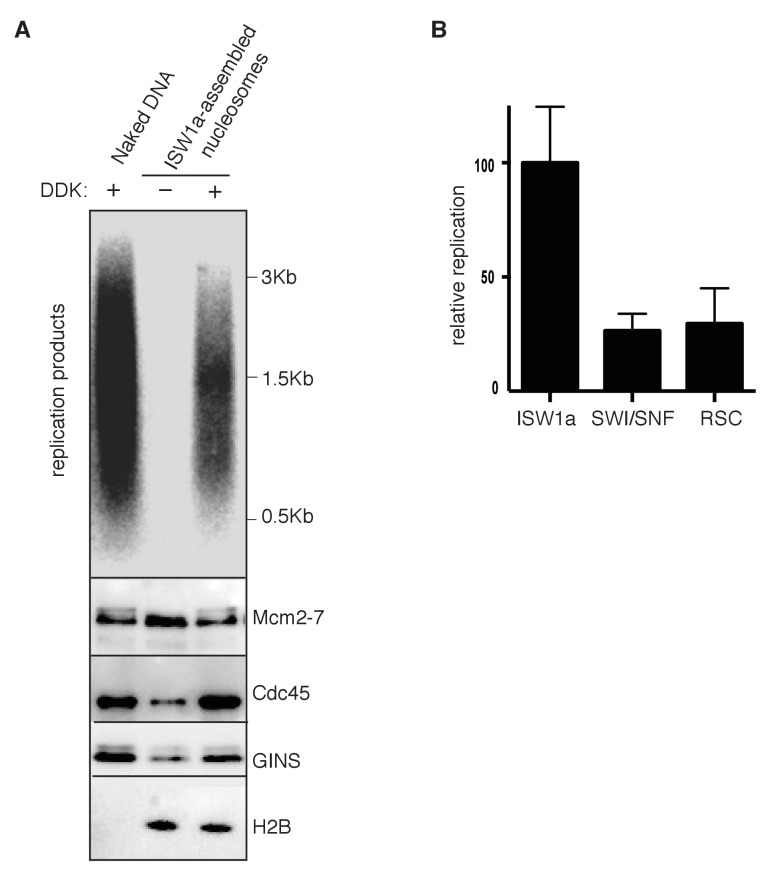
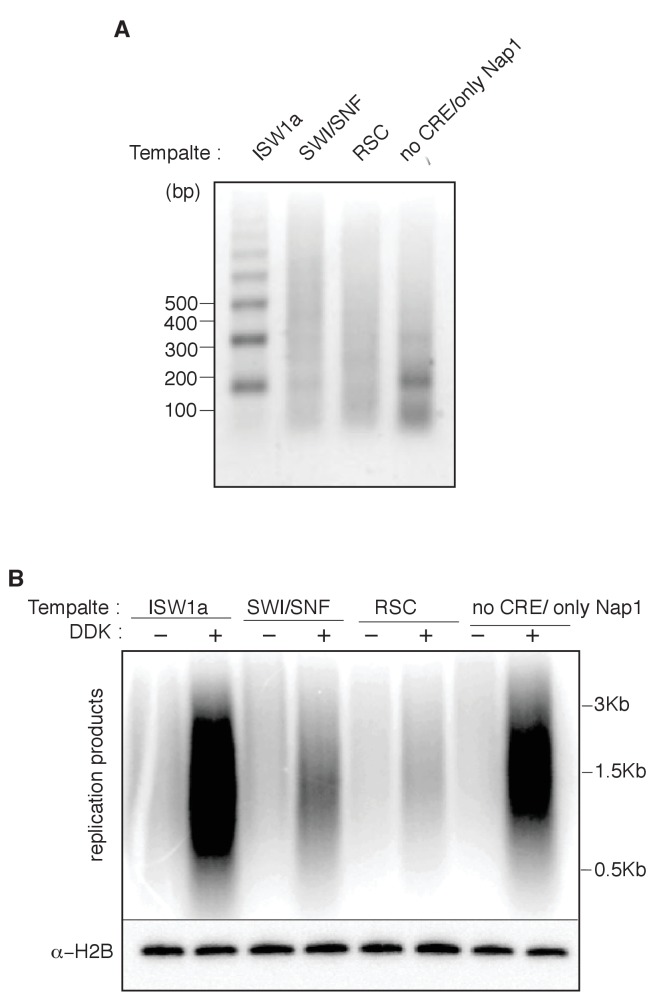
Figure 4. SWI/SNF and RSC templates show reduced CMG formation.
(A) Outline of fully-reconstituted nucleosomal DNA replication initiation assay. The proteins added at each step are indicated. (B) Comparison of reconstituted nucleosomal DNA replication using ISW1a, SWI/SNF and RSC templates in the presence or absence of DDK. Analysis of replication products and H2B as in Figure 3B. (C) Comparison of Mcm2-7 phosphorylation by DDK on ISW1a, SWI/SNF and RSC templates. Phosphorylation of Mcm6 is indicated by reduced electrophoretic mobility and was analyzed by immunoblot (top). Template associated H2B is shown (lower). (D) Comparison of replication of ISW1a, RSC and SWI/SNF templates. Reactions were performed with or without DDK and replication products of the reconstituted replication reactions were analyzed as in Figure 3B (top). Template association of Mcm2-7, Cdc45, GINS and H2B was measured after a high-salt wash at the end of reconstituted replication assay by immunoblot (lower panels). (E) Comparison of CMG formation and activation using ISW1a, SWI/SNF and RSC templates. To prevent replication initiation, the only nucleotide present was ATP and Pol α was left out of the assay. Template association of Mcm2-7, Cdc45, GINS, Rfa1 and H2B were measured by immunoblot.