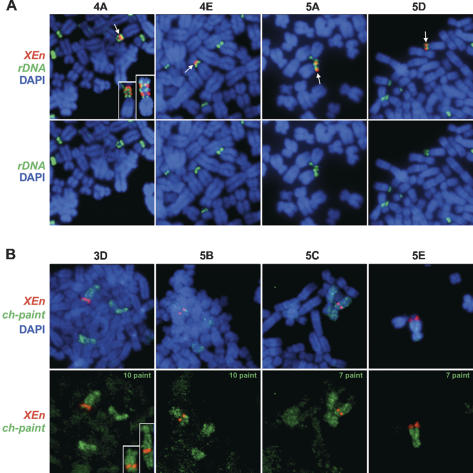
Figure 1.
Generation and mapping of UBF-binding site arrays. (A) XEn arrays map to the short arms of acrocentric chromosomes in clones 4A, 4E, 5A, and 5D. Human NORs were visualized using a spectrum green-labeled probe derived from the intergenic spacer of the human ribosomal gene repeat. XEn sequences were visualized with spectrum red-labeled insert of the plasmid pXEn8. Chromosome identity was determined using enhanced reverse DAPI banding (SmartCapture; data not shown). Merges of both probes and DAPI staining are shown in the top panels. Arrows indicate the XEn arrays. In the bottom panels, only human rDNA and DAPI signals are shown. (B) XEn arrays map to submetacentric chromosomes in clones 3D, 5B, 5C, and 5E. Chromosomes were initially identified using enhanced reverse DAPI banding and then confirmed using chromosome paints labeled with spectrum green. XEn arrays were visualized as above. Merges of both probes and DAPI staining are shown in the top panels. In the bottom panels, DAPI signal is omitted in order to more readily visualize the chromosome paint. The identity of the chromosome paint is shown in the top right corner of each panel.