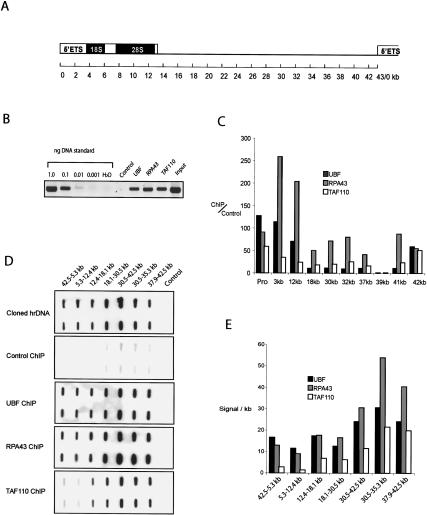
Figure 5.
RNA pol I transcription machinery interacts with DNA sequences across the entire rDNA repeat. (A) The 43-kb human rDNA repeat derived from GenBank accession number U13369 is shown in cartoon form; 0 kb defines the origin of the 13-kb 47S pre rRNA transcript. (B) Nucleolar ChIP. ChIP was performed on nucleolar chromatin prepared from HT1080 cells with affinity-purified UBF, RPA43, and TAF110 antibodies. PCR reactions were performed on one-fortieth of the DNA recovered from each ChIP using primers specific for the human ribosomal gene promoter. A serial dilution of a cosmid comprising the entire human ribosomal repeat was used as a standard template. In a control ChIP, antibody was omitted. (C) Real-time PCR. Real-time PCR was performed on the above ChIPs and DNA standards with promoter specific primers and primers located at a further nine sites across the human rDNA repeat. The amount of target sequence in antibody ChIPs versus control ChIPs was calculated and plotted for each factor and for each primer set. The location of primer pairs is shown below each data set as appropriate. (D) Hybridization with radiolabeled ChIPs. Half of the DNA recovered from antibody and control ChIPs was radiolabeled and used to probe slot blots that were loaded in duplicate with cloned subfragments of the rDNA repeat and vector DNA. To demonstrate uniformity of hybridization across the repeat, the array was also probed with a cosmid clone that comprises the entire 43 kb of human rDNA. The identity of the probe is shown on the left of each panel, and the location of each rDNA subfragment is shown above the appropriate slots. Note that all hybridizations were performed and exposed simultaneously. (E) Quantitation of hybridization. Hybridization signals were quantified by using a Molecular Imager (Bio-Rad). Hybridization signal per kilobase (in arbitrary units) was plotted for each rDNA subfragment and each probe.