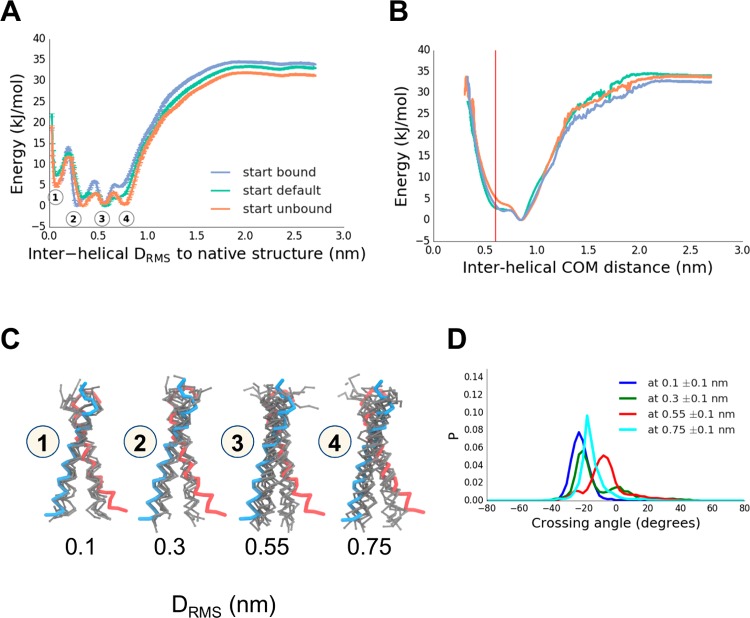
Figure 4.
Dimerization of the TM helix of glycophorin A (GpA), providing an example of a protein–protein interaction within a membrane. (A) GpA dimerization showing a PMF along a reaction coordinate (CV) defined by the intermolecular DRMS to the native dimer structure (PDB id 1AFO). Three initial conditions used, which had either all replicas in the native configuration (“bound”, blue), all replicas initially dissociated (“unbound”, orange), or replicas linearly spaced apart (“default”, cyan). Observed 4 energy minima are labeled 1–4, corresponding to DRMS values of about 0.1, 0.3, 0.55, and 0.75 nm, respectively. (B) Reweighted PMF along a CV corresponding to the interhelical distance (the native structure distance is in red at ∼0.6 nm). (C) Representative structures (in gray; wireframe backbone) from each of the four minima labeled in the PMF in (A). The two subunits of the experimental (PDB id 1AFO) are shown in blue and in red. (D) Crossing angle distributions for the structures seen in PMF minima labeled in (A), with crossing angle defined as a torsion angle between flanking backbone atoms of residues number 78–88.