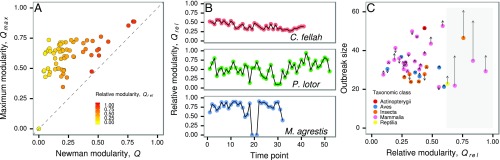
Fig. 2.
Animal social networks with modular subdivisions. (A) Values of Newman modularity, , and maximum possible modularity given the network configuration, , across all animal groups in the database. Point color represents the relative modularity estimated as the ratio of over . The dashed line represents = and the maximum value of . (B) Relative modularity values of dynamic interactions in ants (Camponotus fellah), raccoons (Procyon lotor), and field voles (Microtus agrestis), where time points represent consecutive days, weeks, and trapping sessions, respectively. (C) Comparisons between real (filled points) and their homogeneous null networks (tips of arrows) with respect to the percentage of infected individuals (outbreak size) due to an outbreak with basic reproduction number, . Point color corresponds to the taxonomic class of the animal group. Social networks with nonsignificant modular subdivision (as indicated by t test analysis) have been excluded in C. The generated homogeneous null networks preserve only the local heterogeneity of contacts among individuals; the arrows therefore indicate the change in direction and magnitude of outbreak size under the scenario in which all higher-order structural complexities (including modular subdivisions) are removed from animal social networks. The shaded area represents the region where empirical networks tend to experience reduced outbreak size (at ; SI Appendix, Fig. S2). The networks that experienced lower disease burden included social networks of raccoons (P. lotor), elephant seals (Mirounga angustirostris), ants (Camponotus pennsylvanicus), bottlenose dolphins (Tursiops truncatus), and Australian sleepy lizards (Tiliqua rugosa). In A and C, we used a randomly selected network snapshot for groups with temporal interaction data.