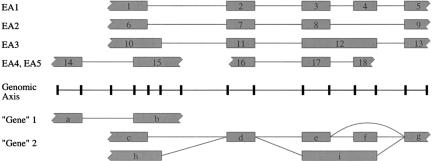
Figure 1.
Splice graph construction from genomic alignments of cDNA and/or protein sequences. A set of representative exon endpoints on the genome (tickmarks) is chosen by reconciling 5′ or 3′ exon endpoints in evidence alignments (EA1-EA5) within 20-bp windows. Exons in the graph (a-i) are produced by enumerating and merging all compatible combinations of evidence exons (1-18), i.e., in which one exon's spliced end is not contained within another exon. (For instance, exons 12 and 17 are incompatible.) Introns are added consistently with the evidence. The graph is refined into connected components (“gene 1” and “gene 2”), representing potentially different overlapping genes. Splice variants can be read from the graph as paths from a node with no incoming edges (source; a, c,h) to a node with no outgoing edges (sink; b, g).