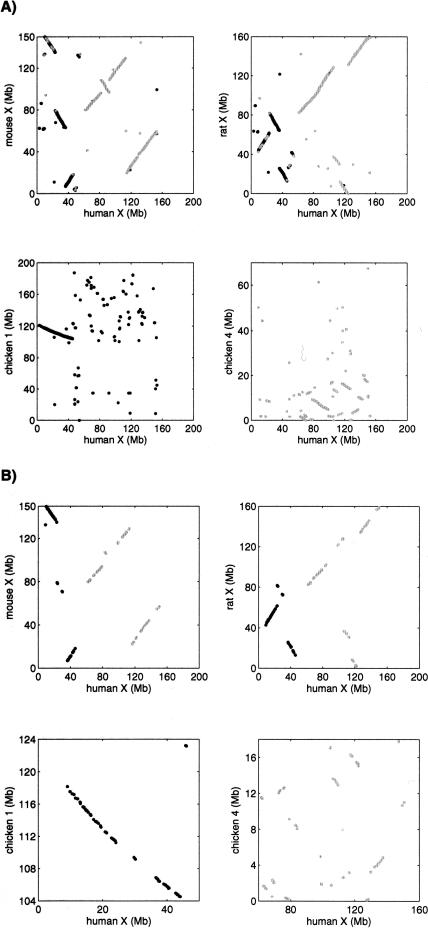
Figure 1.
(A) Dot-plots of anchors based on UCSC alignments for the mammalian X-chromosome versus chicken Chromosomes 1 and 4. The data are very noisy. Many spurious hits are visible on the plots, and there are additional spurious hits between the mammalian X-chromosome and the other chicken chromosomes (data not shown). These spurious hits were filtered out by GRIMM-Synteny because they did not congeal into a block of sufficiently large size. (B) Dot-plots of genes on the mammalian X-chromosome versus chicken Chromosomes 1 and 4. The data are more sparse but much cleaner than the alignment-based data. There were no spurious hits between the mammalian X-chromosome and the other chicken chromosomes.